You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000437_00810
You are here: Home > Sequence: MGYG000000437_00810
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes sp001941065 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; Alistipes sp001941065 | |||||||||||
| CAZyme ID | MGYG000000437_00810 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 22478; End: 23908 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH28 | 141 | 471 | 4.5e-58 | 0.9661538461538461 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5434 | Pgu1 | 1.70e-44 | 106 | 376 | 73 | 382 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| pfam00295 | Glyco_hydro_28 | 1.91e-12 | 207 | 385 | 50 | 234 | Glycosyl hydrolases family 28. Glycosyl hydrolase family 28 includes polygalacturonase EC:3.2.1.15 as well as rhamnogalacturonase A(RGase A), EC:3.2.1.-. These enzymes are important in cell wall metabolism. |
| PLN03003 | PLN03003 | 1.05e-06 | 182 | 357 | 93 | 290 | Probable polygalacturonase At3g15720 |
| TIGR03808 | RR_plus_rpt_1 | 1.66e-05 | 133 | 228 | 54 | 136 | twin-arg-translocated uncharacterized repeat protein. Members of this protein family have a Sec-independent twin-arginine tranlocation (TAT) signal sequence, which enables tranfer of proteins folded around prosthetic groups to cross the plasma membrane. These proteins have four copies of a repeat of about 23 amino acids that resembles the beta-helix repeat. Beta-helix refers to a structural motif in which successive beta strands wind around to stack parallel in a right-handed helix, as in AlgG and related enzymes of carbohydrate metabolism. The twin-arginine motif suggests that members of this protein family bind some unknown cofactor. |
| PLN02218 | PLN02218 | 5.18e-05 | 88 | 476 | 40 | 423 | polygalacturonase ADPG |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADY51374.1 | 8.61e-115 | 2 | 457 | 20 | 474 |
| ADY51373.1 | 1.86e-105 | 1 | 466 | 28 | 506 |
| ADY51372.1 | 1.39e-88 | 7 | 467 | 21 | 496 |
| QIF01889.1 | 1.12e-62 | 25 | 460 | 42 | 486 |
| ADY51377.1 | 2.10e-51 | 23 | 476 | 41 | 507 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5OLP_A | 9.05e-11 | 80 | 347 | 22 | 315 | Galacturonidase[Bacteroides thetaiotaomicron VPI-5482],5OLP_B Galacturonidase [Bacteroides thetaiotaomicron VPI-5482] |
| 2UVE_A | 6.21e-06 | 117 | 385 | 158 | 485 | Structureof Yersinia enterocolitica Family 28 Exopolygalacturonase [Yersinia enterocolitica],2UVE_B Structure of Yersinia enterocolitica Family 28 Exopolygalacturonase [Yersinia enterocolitica],2UVF_A Structure of Yersinia enterocolitica Family 28 Exopolygalacturonase in Complex with Digalaturonic Acid [Yersinia enterocolitica],2UVF_B Structure of Yersinia enterocolitica Family 28 Exopolygalacturonase in Complex with Digalaturonic Acid [Yersinia enterocolitica] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A7PZL3 | 1.14e-16 | 117 | 374 | 64 | 343 | Probable polygalacturonase OS=Vitis vinifera OX=29760 GN=GSVIVT00026920001 PE=1 SV=1 |
| P27644 | 7.99e-11 | 215 | 369 | 4 | 162 | Polygalacturonase OS=Rhizobium radiobacter OX=358 GN=pgl PE=2 SV=1 |
| Q9LW07 | 2.34e-06 | 198 | 357 | 105 | 290 | Probable polygalacturonase At3g15720 OS=Arabidopsis thaliana OX=3702 GN=At3g15720 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
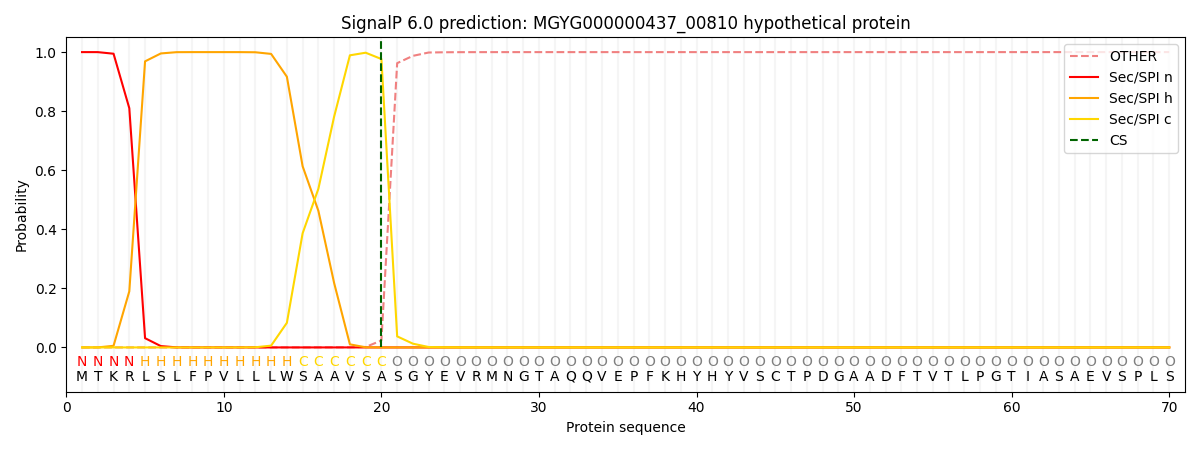
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000267 | 0.999083 | 0.000169 | 0.000156 | 0.000150 | 0.000141 |
