You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000441_00130
You are here: Home > Sequence: MGYG000000441_00130
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-312 sp900545705 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Opitutales; CAG-312; CAG-312; CAG-312 sp900545705 | |||||||||||
| CAZyme ID | MGYG000000441_00130 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 149340; End: 150881 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 89 | 462 | 2.8e-42 | 0.9636963696369637 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00633 | Glyco_10 | 4.60e-32 | 171 | 460 | 17 | 260 | Glycosyl hydrolase family 10. |
| pfam00331 | Glyco_hydro_10 | 5.10e-23 | 173 | 460 | 62 | 305 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 1.26e-21 | 132 | 463 | 45 | 337 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AWI10666.1 | 2.60e-163 | 44 | 513 | 1 | 480 |
| QGA28189.1 | 3.81e-160 | 48 | 513 | 32 | 504 |
| QQZ02681.1 | 8.04e-155 | 37 | 512 | 18 | 494 |
| AHF92621.1 | 1.28e-153 | 42 | 503 | 16 | 481 |
| AVM47074.1 | 6.04e-148 | 31 | 513 | 5 | 496 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1NQ6_A | 1.24e-08 | 171 | 460 | 62 | 295 | CrystalStructure of the catalytic domain of xylanase A from Streptomyces halstedii JM8 [Streptomyces halstedii] |
| 4W8L_A | 2.72e-06 | 172 | 463 | 64 | 341 | Structureof GH10 from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4W8L_B Structure of GH10 from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4W8L_C Structure of GH10 from Paenibacillus barcinonensis [Paenibacillus barcinonensis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P49942 | 2.30e-07 | 249 | 459 | 131 | 364 | Endo-1,4-beta-xylanase A OS=Bacteroides ovatus OX=28116 GN=xylI PE=2 SV=1 |
| A0A1P8AWH8 | 1.23e-06 | 58 | 496 | 562 | 913 | Endo-1,4-beta-xylanase 1 OS=Arabidopsis thaliana OX=3702 GN=XYN1 PE=1 SV=1 |
| O69230 | 2.92e-06 | 172 | 498 | 430 | 740 | Endo-1,4-beta-xylanase C OS=Paenibacillus barcinonensis OX=198119 GN=xynC PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
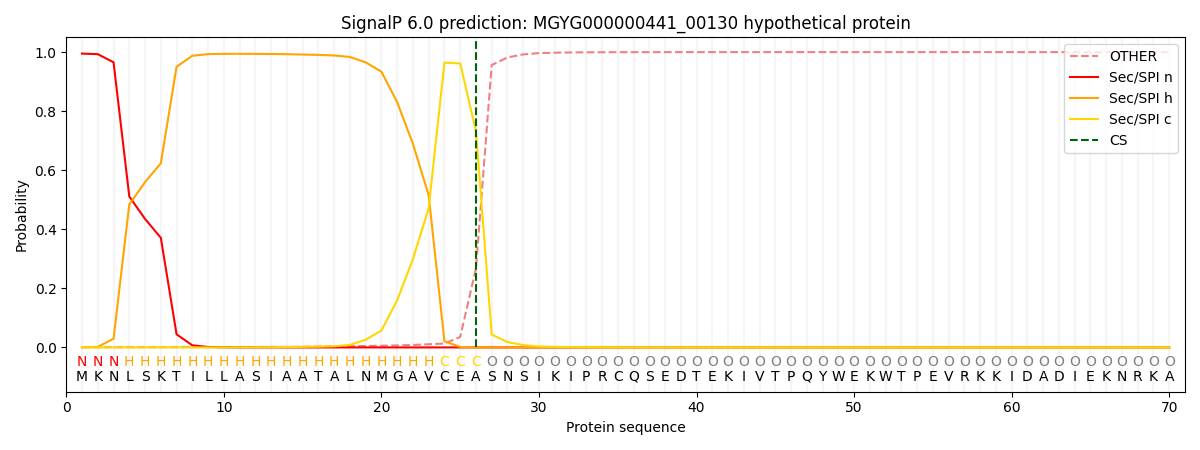
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002053 | 0.992191 | 0.004885 | 0.000389 | 0.000240 | 0.000209 |
