You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000441_00643
You are here: Home > Sequence: MGYG000000441_00643
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-312 sp900545705 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Opitutales; CAG-312; CAG-312; CAG-312 sp900545705 | |||||||||||
| CAZyme ID | MGYG000000441_00643 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | Anti-sigma-I factor RsgI6 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 7928; End: 9424 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 131 | 435 | 1.9e-37 | 0.8943894389438944 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00633 | Glyco_10 | 4.18e-28 | 149 | 413 | 3 | 238 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 3.55e-24 | 149 | 412 | 69 | 305 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam00331 | Glyco_hydro_10 | 1.50e-18 | 149 | 413 | 46 | 279 | Glycosyl hydrolase family 10. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QMW04636.1 | 6.54e-120 | 47 | 489 | 20 | 450 |
| QHV94149.1 | 2.25e-99 | 56 | 493 | 26 | 442 |
| QIP12711.1 | 2.51e-98 | 56 | 493 | 26 | 442 |
| AEI51978.1 | 5.08e-97 | 56 | 493 | 26 | 438 |
| QMW04637.1 | 3.20e-96 | 56 | 488 | 26 | 437 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7D88_A | 3.56e-21 | 58 | 467 | 41 | 393 | ChainA, Beta-xylanase [Bacillus sp. (in: Bacteria)] |
| 7D89_A | 1.22e-19 | 58 | 467 | 41 | 393 | ChainA, Beta-xylanase [Bacillus sp. (in: Bacteria)] |
| 1TA3_B | 1.59e-08 | 177 | 330 | 66 | 209 | CrystalStructure of xylanase (GH10) in complex with inhibitor (XIP) [Aspergillus nidulans] |
| 3EMC_A | 1.39e-07 | 177 | 411 | 64 | 294 | Crystalstructure of XynB, an intracellular xylanase from Paenibacillus barcinonensis [Paenibacillus barcinonensis],3EMQ_A Crystal structure of xilanase XynB from Paenibacillus barcelonensis complexed with an inhibitor [Paenibacillus barcinonensis],3EMZ_A Crystal structure of xylanase XynB from Paenibacillus barcinonensis complexed with a conduramine derivative [Paenibacillus barcinonensis] |
| 1XAS_A | 2.09e-06 | 152 | 338 | 52 | 216 | CRYSTALSTRUCTURE, AT 2.6 ANGSTROMS RESOLUTION, OF THE STREPTOMYCES LIVIDANS XYLANASE A, A MEMBER OF THE F FAMILY OF BETA-1,4-D-GLYCANSES [Streptomyces lividans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A3DH97 | 7.34e-19 | 55 | 485 | 379 | 747 | Anti-sigma-I factor RsgI6 OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=rsgI6 PE=1 SV=1 |
| A0A1P8B8F8 | 1.18e-09 | 149 | 467 | 250 | 533 | Endo-1,4-beta-xylanase 5 OS=Arabidopsis thaliana OX=3702 GN=XYN5 PE=3 SV=1 |
| Q84WT5 | 2.07e-09 | 177 | 467 | 266 | 534 | Endo-1,4-beta-xylanase 5-like OS=Arabidopsis thaliana OX=3702 GN=At4g33820 PE=2 SV=1 |
| P40944 | 7.06e-09 | 177 | 369 | 415 | 599 | Endo-1,4-beta-xylanase A OS=Caldicellulosiruptor sp. (strain Rt8B.4) OX=28238 GN=xynA PE=3 SV=1 |
| Q00177 | 1.00e-07 | 177 | 330 | 90 | 233 | Endo-1,4-beta-xylanase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=xlnC PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
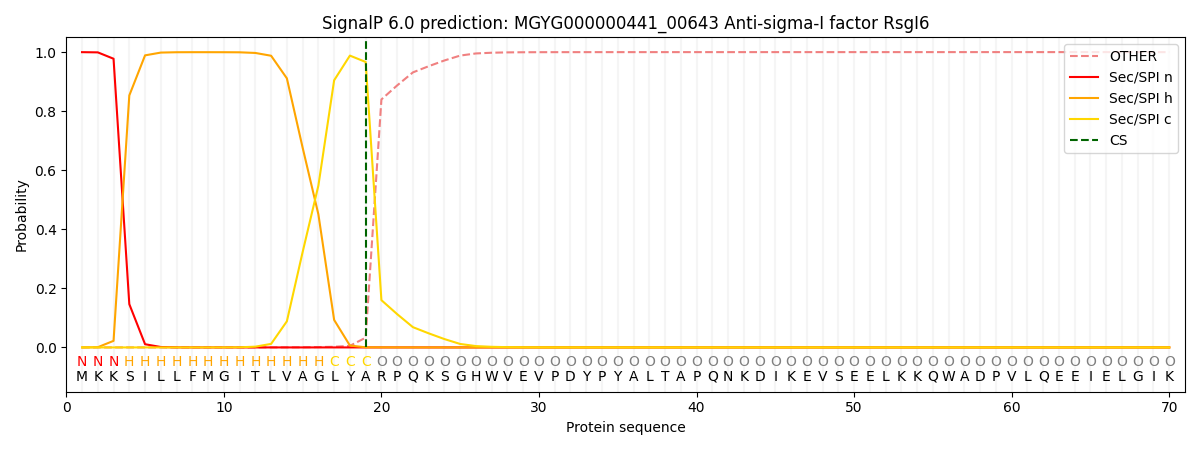
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000754 | 0.998138 | 0.000517 | 0.000197 | 0.000184 | 0.000176 |
