You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000442_02253
You are here: Home > Sequence: MGYG000000442_02253
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-462 sp003489705 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; CAG-462; CAG-462 sp003489705 | |||||||||||
| CAZyme ID | MGYG000000442_02253 | |||||||||||
| CAZy Family | PL10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 13440; End: 14534 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL10 | 60 | 348 | 6.4e-116 | 0.9930555555555556 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam09492 | Pec_lyase | 1.33e-147 | 60 | 350 | 1 | 289 | Pectic acid lyase. Members of this family are isozymes of pectate lyase (EC:4.2.2.2), also called polygalacturonic transeliminase and alpha-1,4-D-endopolygalacturonic acid lyase. |
| TIGR02474 | pec_lyase | 1.06e-132 | 60 | 350 | 1 | 290 | pectate lyase, PelA/Pel-15E family. Members of this family are isozymes of pectate lyase (EC 4.2.2.2), also called polygalacturonic transeliminase and alpha-1,4-D-endopolygalacturonic acid lyase. [Energy metabolism, Biosynthesis and degradation of polysaccharides] |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ALJ01761.1 | 5.06e-127 | 26 | 359 | 34 | 372 |
| AKP52260.1 | 1.35e-126 | 2 | 359 | 9 | 370 |
| QEC53563.1 | 1.60e-125 | 30 | 356 | 38 | 365 |
| AEL26628.1 | 1.25e-124 | 9 | 359 | 13 | 370 |
| SOE21140.1 | 1.19e-123 | 4 | 361 | 2 | 358 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1R76_A | 4.96e-69 | 33 | 359 | 63 | 407 | ChainA, pectate lyase [Niveispirillum irakense] |
| 1GXM_A | 1.28e-65 | 37 | 350 | 26 | 323 | Family10 polysaccharide lyase from Cellvibrio cellulosa [Cellvibrio japonicus],1GXM_B Family 10 polysaccharide lyase from Cellvibrio cellulosa [Cellvibrio japonicus],1GXN_A Family 10 polysaccharide lyase from Cellvibrio cellulosa [Cellvibrio japonicus] |
| 1GXO_A | 1.99e-64 | 37 | 350 | 26 | 323 | MutantD189A of Family 10 polysaccharide lyase from Cellvibrio cellulosa in complex with trigalaturonic acid [Cellvibrio japonicus] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
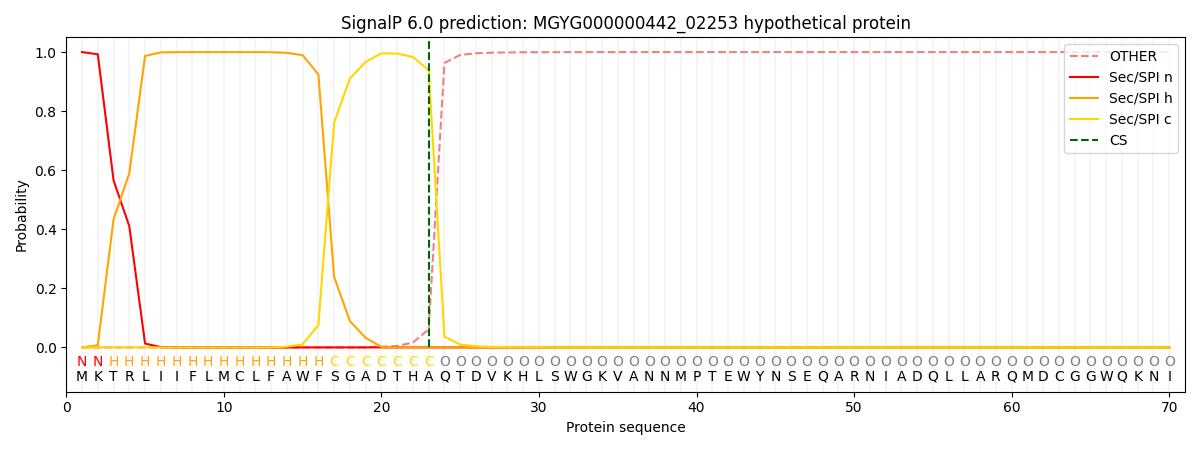
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000274 | 0.999098 | 0.000196 | 0.000149 | 0.000133 | 0.000128 |
