You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000442_02444
You are here: Home > Sequence: MGYG000000442_02444
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-462 sp003489705 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; CAG-462; CAG-462 sp003489705 | |||||||||||
| CAZyme ID | MGYG000000442_02444 | |||||||||||
| CAZy Family | CE19 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 16440; End: 18410 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE19 | 119 | 447 | 1.7e-136 | 0.9849397590361446 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam12875 | DUF3826 | 2.46e-88 | 463 | 644 | 1 | 185 | Protein of unknown function (DUF3826). This is a putative sugar-binding family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRN01959.1 | 0.0 | 3 | 650 | 2 | 651 |
| QUT29074.1 | 0.0 | 3 | 650 | 2 | 651 |
| QDH55640.1 | 0.0 | 3 | 650 | 2 | 651 |
| QUT23580.1 | 0.0 | 3 | 650 | 2 | 651 |
| CBK65855.1 | 0.0 | 9 | 650 | 2 | 644 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6GOC_A | 1.11e-313 | 18 | 648 | 22 | 654 | ChainA, DUF3826 domain-containing protein [Bacteroides thetaiotaomicron] |
| 3KDW_A | 2.29e-12 | 458 | 629 | 28 | 200 | ChainA, Putative sugar binding protein [Phocaeicola vulgatus ATCC 8482] |
| 3G6I_A | 6.67e-08 | 464 | 631 | 16 | 185 | Crystalstructure of an outer membrane protein, part of a putative carbohydrate binding complex (bt_1022) from bacteroides thetaiotaomicron vpi-5482 at 1.93 A resolution [Bacteroides thetaiotaomicron VPI-5482] |
| 6AGQ_A | 3.43e-06 | 154 | 312 | 39 | 214 | Acetylxylan esterase from Paenibacillus sp. R4 [Paenibacillus sp. R4],6AGQ_B Acetyl xylan esterase from Paenibacillus sp. R4 [Paenibacillus sp. R4],6AGQ_C Acetyl xylan esterase from Paenibacillus sp. R4 [Paenibacillus sp. R4],6AGQ_D Acetyl xylan esterase from Paenibacillus sp. R4 [Paenibacillus sp. R4],6AGQ_E Acetyl xylan esterase from Paenibacillus sp. R4 [Paenibacillus sp. R4],6AGQ_F Acetyl xylan esterase from Paenibacillus sp. R4 [Paenibacillus sp. R4] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
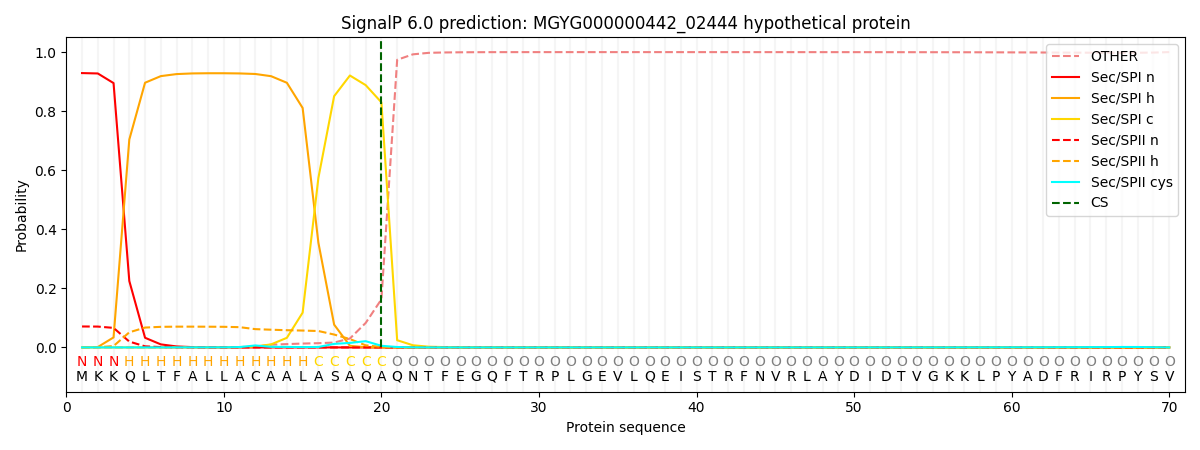
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001059 | 0.919700 | 0.078125 | 0.000431 | 0.000338 | 0.000321 |
