You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000448_00842
You are here: Home > Sequence: MGYG000000448_00842
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-873; | |||||||||||
| CAZyme ID | MGYG000000448_00842 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 6734; End: 8848 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 443 | 698 | 5.6e-48 | 0.6798679867986799 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00633 | Glyco_10 | 3.87e-37 | 443 | 696 | 54 | 263 | Glycosyl hydrolase family 10. |
| pfam00331 | Glyco_hydro_10 | 7.35e-37 | 447 | 698 | 100 | 310 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 2.21e-22 | 441 | 701 | 117 | 342 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam00331 | Glyco_hydro_10 | 1.99e-07 | 85 | 135 | 35 | 85 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 2.48e-05 | 94 | 136 | 1 | 43 | Glycosyl hydrolase family 10. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCD43725.1 | 4.26e-297 | 178 | 703 | 1 | 527 |
| ALJ61518.1 | 8.74e-282 | 1 | 703 | 1 | 721 |
| ADE81777.1 | 2.42e-214 | 2 | 703 | 3 | 707 |
| QVJ82141.1 | 1.95e-213 | 2 | 703 | 3 | 707 |
| AAB81559.1 | 3.90e-213 | 2 | 703 | 3 | 707 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4XUY_A | 4.07e-17 | 442 | 700 | 101 | 302 | Crystalstructure of an endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Aspergillus niger [Aspergillus niger CBS 513.88],4XUY_B Crystal structure of an endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Aspergillus niger [Aspergillus niger CBS 513.88] |
| 1GOK_A | 8.08e-16 | 451 | 700 | 108 | 301 | Thermostablexylanase I from Thermoascus aurantiacus- Crystal form II [Thermoascus aurantiacus],1GOM_A Thermostable xylanase I from Thermoascus aurantiacus- Crystal form I [Thermoascus aurantiacus],1GOO_A Thermostable xylanase I from Thermoascus aurantiacus - Cryocooled glycerol complex [Thermoascus aurantiacus],1GOQ_A Thermostable xylanase I from Thermoascus aurantiacus - Room temperature xylobiose complex [Thermoascus aurantiacus],1GOR_A THERMOSTABLE XYLANASE I FROM THERMOASCUS AURANTIACUS - XYLOBIOSE COMPLEX AT 100 K [Thermoascus aurantiacus] |
| 1K6A_A | 8.08e-16 | 451 | 700 | 108 | 301 | Structuralstudies on the mobility in the active site of the Thermoascus aurantiacus xylanase I [Thermoascus aurantiacus] |
| 3W24_A | 2.91e-15 | 469 | 698 | 141 | 325 | Thehigh-resolution crystal structure of TsXylA, intracellular xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485 [Thermoanaerobacterium saccharolyticum JW/SL-YS485] |
| 1I1W_A | 3.56e-15 | 451 | 700 | 108 | 301 | 0.89AUltra high resolution structure of a Thermostable Xylanase from Thermoascus Aurantiacus [Thermoascus aurantiacus],1I1X_A 1.11 A ATOMIC RESOLUTION STRUCTURE OF A THERMOSTABLE XYLANASE FROM THERMOASCUS AURANTIACUS [Thermoascus aurantiacus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q4P902 | 6.55e-17 | 464 | 700 | 156 | 339 | Endo-1,4-beta-xylanase UM03411 OS=Ustilago maydis (strain 521 / FGSC 9021) OX=237631 GN=UMAG_03411 PE=1 SV=1 |
| P33559 | 2.32e-16 | 442 | 700 | 126 | 327 | Endo-1,4-beta-xylanase A OS=Aspergillus kawachii (strain NBRC 4308) OX=1033177 GN=xynA PE=1 SV=2 |
| A2QFV7 | 3.12e-16 | 442 | 700 | 126 | 327 | Probable endo-1,4-beta-xylanase C OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=xlnC PE=1 SV=1 |
| C5J411 | 4.20e-16 | 442 | 700 | 126 | 327 | Probable endo-1,4-beta-xylanase C OS=Aspergillus niger OX=5061 GN=xlnC PE=2 SV=2 |
| P23360 | 6.11e-15 | 451 | 700 | 134 | 327 | Endo-1,4-beta-xylanase OS=Thermoascus aurantiacus OX=5087 GN=XYNA PE=1 SV=4 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
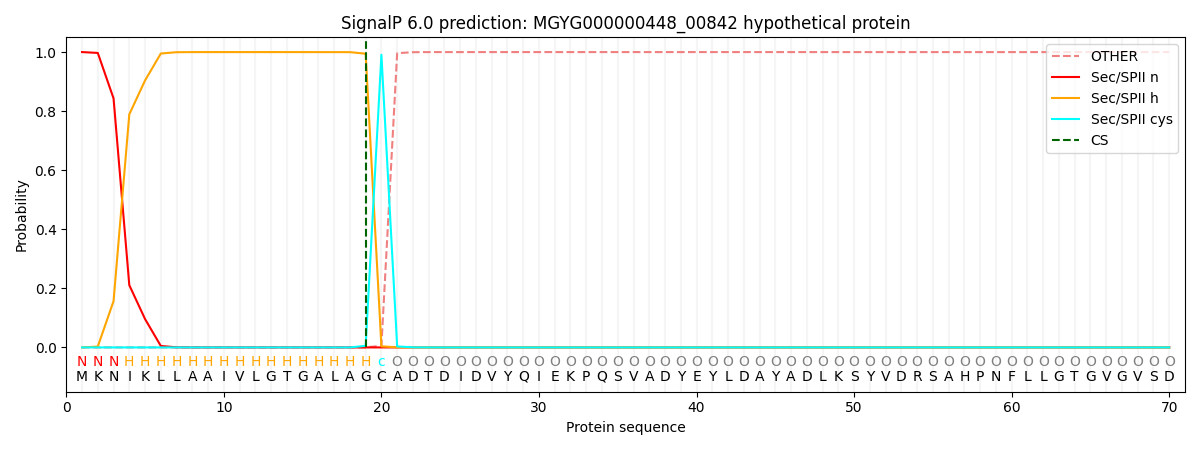
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000001 | 1.000044 | 0.000000 | 0.000000 | 0.000000 |
