You are browsing environment: HUMAN GUT
MGYG000000451_00512
Basic Information
help
Species
UBA1820 sp002314265
Lineage
Bacteria; Bacteroidota; Bacteroidia; Flavobacteriales; UBA1820; UBA1820; UBA1820 sp002314265
CAZyme ID
MGYG000000451_00512
CAZy Family
GT83
CAZyme Description
Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase
CAZyme Property
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000000451
1842661
MAG
Sweden
Europe
Gene Location
Start: 67941;
End: 69626
Strand: +
No EC number prediction in MGYG000000451_00512.
Family
Start
End
Evalue
family coverage
GT83
32
461
5.9e-64
0.8092592592592592
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
COG1807
ArnT
6.11e-24
40
441
23
422
4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis].
more
PRK13279
arnT
1.02e-08
49
442
31
430
lipid IV(A) 4-amino-4-deoxy-L-arabinosyltransferase.
more
pfam13231
PMT_2
1.03e-06
80
236
1
151
Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366.
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
5EZM_A
1.10e-09
44
359
51
372
CrystalStructure of ArnT from Cupriavidus metallidurans in the apo state [Cupriavidus metallidurans CH34],5F15_A Crystal Structure of ArnT from Cupriavidus metallidurans bound to Undecaprenyl phosphate [Cupriavidus metallidurans CH34]
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
O67270
2.16e-13
34
358
5
328
Uncharacterized protein aq_1220 OS=Aquifex aeolicus (strain VF5) OX=224324 GN=aq_1220 PE=3 SV=1
more
A8GDR9
8.30e-10
47
385
30
371
Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Serratia proteamaculans (strain 568) OX=399741 GN=arnT PE=3 SV=1
more
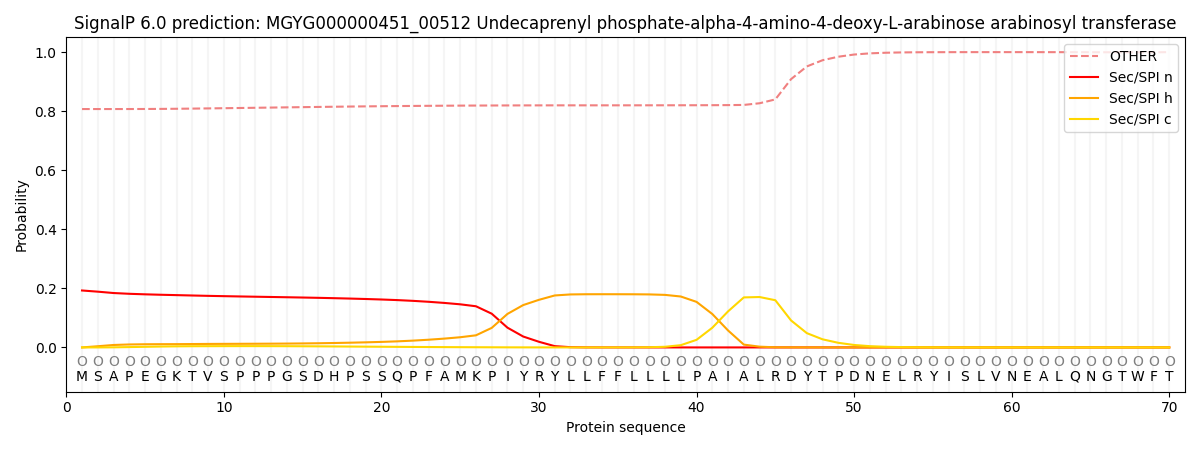
This protein is predicted as OTHER
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.813283
0.186027
0.000222
0.000123
0.000128
0.000225
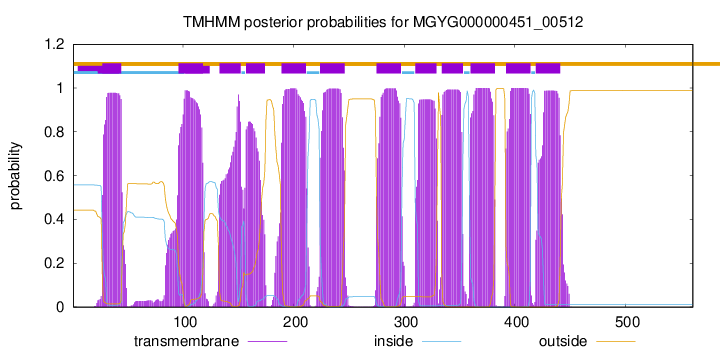
start
end
27
44
96
118
133
152
157
174
189
211
224
246
275
297
310
329
334
353
360
382
392
414
419
441