You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000462_00416
You are here: Home > Sequence: MGYG000000462_00416
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UBA940 sp900768115 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; UBA940; UBA940 sp900768115 | |||||||||||
| CAZyme ID | MGYG000000462_00416 | |||||||||||
| CAZy Family | GH109 | |||||||||||
| CAZyme Description | Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 49184; End: 50395 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH109 | 34 | 199 | 1e-24 | 0.39598997493734334 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0673 | MviM | 8.18e-35 | 32 | 386 | 2 | 342 | Predicted dehydrogenase [General function prediction only]. |
| pfam01408 | GFO_IDH_MocA | 2.14e-11 | 34 | 158 | 1 | 110 | Oxidoreductase family, NAD-binding Rossmann fold. This family of enzymes utilize NADP or NAD. This family is called the GFO/IDH/MOCA family in swiss-prot. |
| COG4091 | COG4091 | 8.28e-05 | 23 | 137 | 7 | 129 | Predicted homoserine dehydrogenase, contains C-terminal SAF domain [Amino acid transport and metabolism]. |
| pfam02894 | GFO_IDH_MocA_C | 2.18e-04 | 179 | 314 | 2 | 116 | Oxidoreductase family, C-terminal alpha/beta domain. This family of enzymes utilize NADP or NAD. This family is called the GFO/IDH/MOCA family in swiss-prot. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBD45948.1 | 2.05e-141 | 3 | 403 | 6 | 423 |
| QEC68724.1 | 1.19e-132 | 2 | 403 | 5 | 423 |
| BBD45631.1 | 1.50e-127 | 1 | 403 | 1 | 420 |
| SCD19022.1 | 1.72e-126 | 1 | 403 | 1 | 420 |
| CEA16880.1 | 2.56e-123 | 1 | 403 | 1 | 420 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3EZY_A | 5.01e-14 | 34 | 259 | 3 | 197 | Crystalstructure of probable dehydrogenase TM_0414 from Thermotoga maritima [Thermotoga maritima],3EZY_B Crystal structure of probable dehydrogenase TM_0414 from Thermotoga maritima [Thermotoga maritima],3EZY_C Crystal structure of probable dehydrogenase TM_0414 from Thermotoga maritima [Thermotoga maritima],3EZY_D Crystal structure of probable dehydrogenase TM_0414 from Thermotoga maritima [Thermotoga maritima] |
| 3NT2_A | 2.94e-13 | 34 | 193 | 3 | 149 | Crystalstructure of myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor [Bacillus subtilis],3NT2_B Crystal structure of myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor [Bacillus subtilis],3NT4_A Crystal structure of myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor NADH and inositol [Bacillus subtilis],3NT4_B Crystal structure of myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor NADH and inositol [Bacillus subtilis],3NT5_A Crystal structure of myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor and product inosose [Bacillus subtilis],3NT5_B Crystal structure of myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor and product inosose [Bacillus subtilis] |
| 3MZ0_A | 3.94e-13 | 34 | 193 | 3 | 149 | Crystalstructure of apo myo-inositol dehydrogenase from Bacillus subtilis [Bacillus subtilis] |
| 4L8V_A | 2.19e-12 | 34 | 193 | 3 | 149 | CrystalStructure of A12K/D35S mutant myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor NADP [Bacillus subtilis subsp. subtilis str. 168],4L8V_B Crystal Structure of A12K/D35S mutant myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor NADP [Bacillus subtilis subsp. subtilis str. 168],4L8V_C Crystal Structure of A12K/D35S mutant myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor NADP [Bacillus subtilis subsp. subtilis str. 168],4L8V_D Crystal Structure of A12K/D35S mutant myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor NADP [Bacillus subtilis subsp. subtilis str. 168] |
| 4L9R_A | 2.19e-12 | 34 | 193 | 3 | 149 | CrystalStructure of apo A12K/D35S mutant myo-inositol dehydrogenase from Bacillus subtilis [Bacillus subtilis subsp. subtilis str. 168] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O05389 | 7.59e-15 | 37 | 316 | 8 | 257 | Uncharacterized oxidoreductase YrbE OS=Bacillus subtilis (strain 168) OX=224308 GN=yrbE PE=3 SV=2 |
| Q9WYP5 | 4.59e-13 | 34 | 259 | 1 | 195 | Myo-inositol 2-dehydrogenase OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=iolG PE=1 SV=1 |
| P26935 | 1.61e-12 | 34 | 193 | 3 | 149 | Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase OS=Bacillus subtilis (strain 168) OX=224308 GN=iolG PE=1 SV=2 |
| A0KV43 | 4.70e-11 | 1 | 199 | 4 | 213 | Glycosyl hydrolase family 109 protein 1 OS=Shewanella sp. (strain ANA-3) OX=94122 GN=Shewana3_1428 PE=3 SV=1 |
| A6WQ58 | 6.26e-11 | 1 | 199 | 4 | 213 | Glycosyl hydrolase family 109 protein OS=Shewanella baltica (strain OS185) OX=402882 GN=Shew185_2813 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as TAT
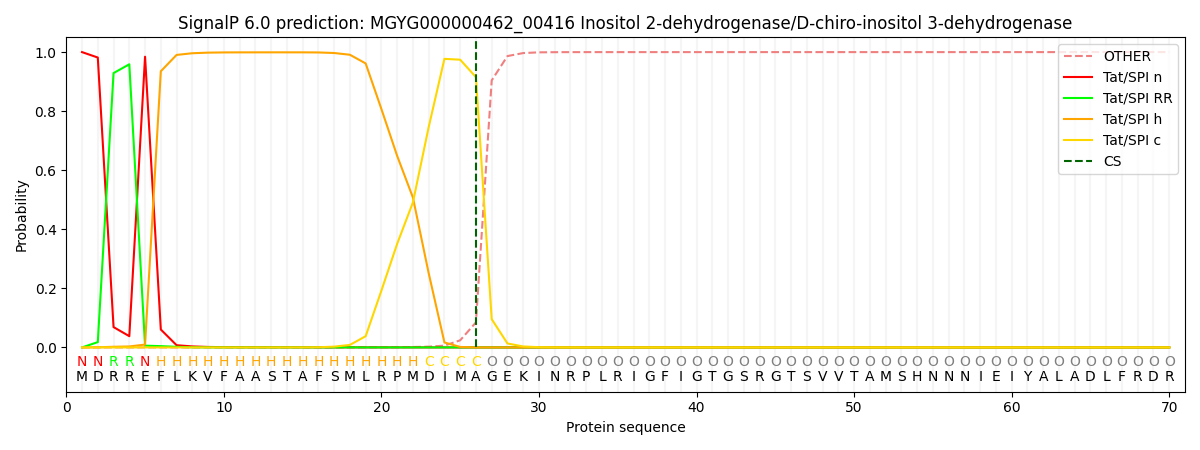
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 0.000000 | 0.999987 | 0.000000 | 0.000000 |
