You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000470_02090
You are here: Home > Sequence: MGYG000000470_02090
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Brachyspira pilosicoli | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Spirochaetota; Brachyspirae; Brachyspirales; Brachyspiraceae; Brachyspira; Brachyspira pilosicoli | |||||||||||
| CAZyme ID | MGYG000000470_02090 | |||||||||||
| CAZy Family | GT76 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1035; End: 2183 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT76 | 66 | 370 | 2.5e-16 | 0.7027027027027027 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5542 | COG5542 | 2.88e-04 | 62 | 271 | 78 | 281 | Mannosyltransferase related to Gpi18 [Carbohydrate transport and metabolism]. |
| pfam04188 | Mannosyl_trans2 | 0.005 | 62 | 192 | 61 | 203 | Mannosyltransferase (PIG-V). This is a family of eukaryotic ER membrane proteins that are involved in the synthesis of glycosylphosphatidylinositol (GPI), a glycolipid that anchors many proteins to the eukaryotic cell surface. Proteins in this family are involved in transferring the second mannose in the biosynthetic pathway of GPI. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AGA67034.1 | 2.72e-262 | 1 | 382 | 1 | 382 |
| QGZ92645.1 | 9.71e-16 | 54 | 355 | 64 | 367 |
| BBH39378.1 | 1.05e-14 | 54 | 355 | 64 | 367 |
| AKM83719.1 | 3.80e-14 | 62 | 351 | 58 | 331 |
| SNX53584.1 | 9.40e-14 | 62 | 353 | 80 | 388 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000040 | 0.000001 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
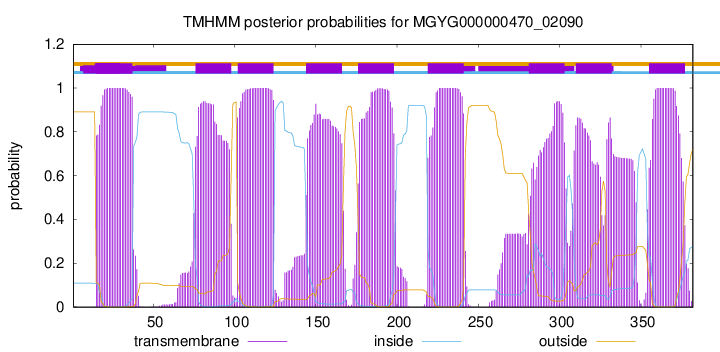
| start | end |
|---|---|
| 15 | 37 |
| 76 | 98 |
| 102 | 124 |
| 144 | 166 |
| 176 | 198 |
| 219 | 241 |
| 281 | 303 |
| 310 | 332 |
| 355 | 377 |
