You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000470_02255
You are here: Home > Sequence: MGYG000000470_02255
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Brachyspira pilosicoli | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Spirochaetota; Brachyspirae; Brachyspirales; Brachyspiraceae; Brachyspira; Brachyspira pilosicoli | |||||||||||
| CAZyme ID | MGYG000000470_02255 | |||||||||||
| CAZy Family | CBM50 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2571; End: 3704 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| NF033163 | lipo_LipL71 | 9.29e-15 | 297 | 360 | 405 | 467 | lipoprotein LipL71. Members of this family are lipoprotein LipL71, also known as LruA, as described in Leptospira interrogans but found broadly in the genus Leptospira. Close homologs that are not lipoproteins by sequence are likely defective in their reported coding region. |
| PRK11198 | PRK11198 | 9.38e-10 | 296 | 353 | 95 | 146 | LysM domain/BON superfamily protein; Provisional |
| COG1652 | XkdP | 6.74e-07 | 301 | 360 | 211 | 269 | Nucleoid-associated protein YgaU, contains BON and LysM domains [Function unknown]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AFR70188.1 | 7.64e-271 | 1 | 377 | 1 | 377 |
| CCG57295.1 | 7.64e-271 | 1 | 377 | 1 | 377 |
| AGA65437.1 | 6.27e-270 | 1 | 377 | 1 | 377 |
| AEM22701.1 | 2.05e-134 | 26 | 373 | 22 | 371 |
| ASJ20730.1 | 2.19e-134 | 1 | 373 | 1 | 388 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P19649 | 3.26e-14 | 292 | 370 | 240 | 321 | Treponemal membrane protein B OS=Treponema pallidum (strain Nichols) OX=243276 GN=tmpB PE=3 SV=2 |
| P29720 | 3.13e-11 | 294 | 370 | 299 | 378 | Treponemal membrane protein B OS=Treponema phagedenis OX=162 GN=tmpB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
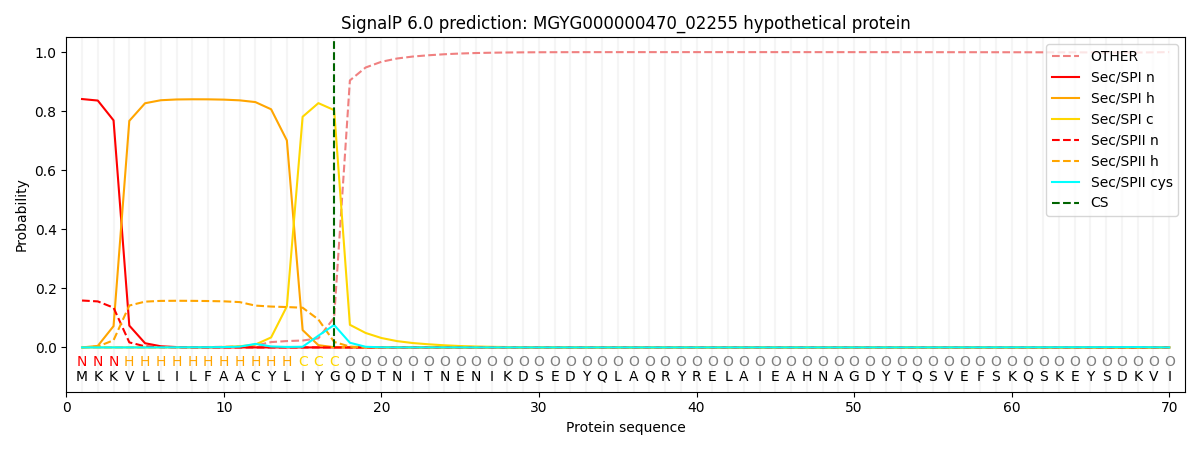
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000893 | 0.832147 | 0.166182 | 0.000250 | 0.000251 | 0.000238 |
