You are browsing environment: HUMAN GUT
MGYG000000480_01353
Basic Information
help
Species
CAG-312 sp900546565
Lineage
Bacteria; Verrucomicrobiota; Verrucomicrobiae; Opitutales; CAG-312; CAG-312; CAG-312 sp900546565
CAZyme ID
MGYG000000480_01353
CAZy Family
GH28
CAZyme Description
hypothetical protein
CAZyme Property
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000000480
1940095
MAG
Fiji
Oceania
Gene Location
Start: 4099;
End: 5253
Strand: +
No EC number prediction in MGYG000000480_01353.
Family
Start
End
Evalue
family coverage
GH28
118
350
3.7e-21
0.6615384615384615
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
COG5434
Pgu1
4.94e-10
39
348
97
402
Polygalacturonase [Carbohydrate transport and metabolism].
more
pfam00295
Glyco_hydro_28
5.58e-08
121
269
48
204
Glycosyl hydrolases family 28. Glycosyl hydrolase family 28 includes polygalacturonase EC:3.2.1.15 as well as rhamnogalacturonase A(RGase A), EC:3.2.1.-. These enzymes are important in cell wall metabolism.
more
pfam13229
Beta_helix
7.58e-04
153
281
2
119
Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases.
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
AWO01000.1
9.80e-118
31
362
5
331
APZ82864.1
2.40e-30
31
362
26
339
BBZ75998.1
5.12e-06
153
305
159
287
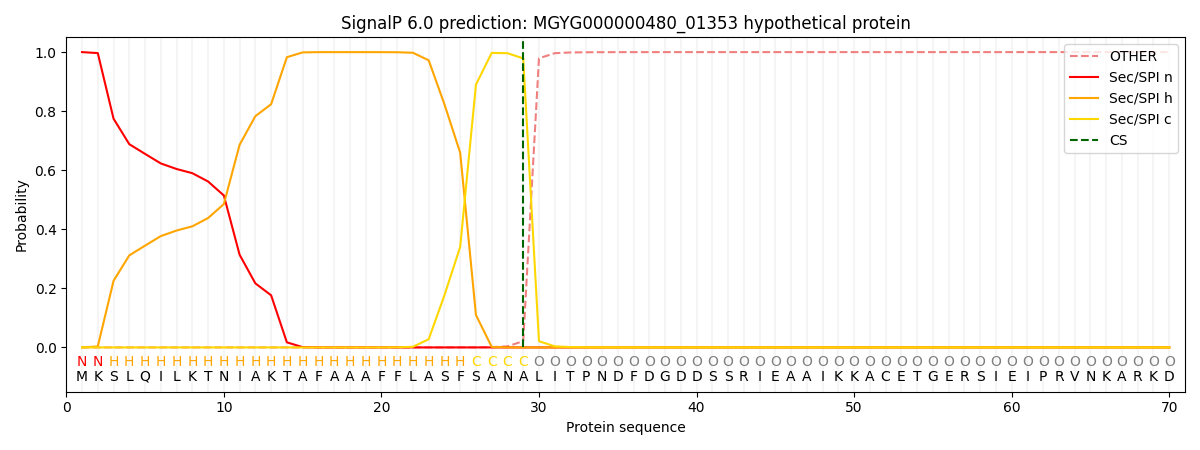
This protein is predicted as SP
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.000263
0.999092
0.000158
0.000169
0.000148
0.000136
There is no transmembrane helices in MGYG000000480_01353.