You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000484_01548
You are here: Home > Sequence: MGYG000000484_01548
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
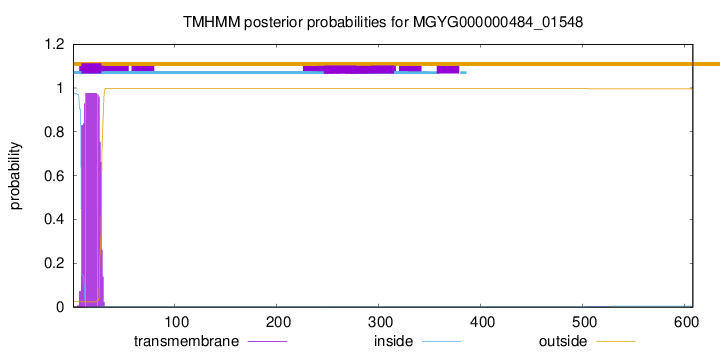
TMHMM annotations
Basic Information help
| Species | Butyrivibrio_A sp000431815 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Butyrivibrio_A; Butyrivibrio_A sp000431815 | |||||||||||
| CAZyme ID | MGYG000000484_01548 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 39421; End: 41247 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 96 | 386 | 8.4e-86 | 0.9927536231884058 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 2.67e-43 | 87 | 351 | 6 | 239 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 7.70e-28 | 45 | 427 | 27 | 407 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| pfam01301 | Glyco_hydro_35 | 4.11e-07 | 107 | 211 | 27 | 143 | Glycosyl hydrolases family 35. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| VEU81114.1 | 1.47e-204 | 61 | 605 | 32 | 576 |
| CBK96866.1 | 7.04e-194 | 60 | 607 | 38 | 584 |
| CCO05195.1 | 3.32e-192 | 46 | 604 | 39 | 591 |
| AHF25528.1 | 1.09e-188 | 58 | 604 | 11 | 573 |
| QTE66996.1 | 3.06e-184 | 58 | 604 | 26 | 588 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3ZMR_A | 4.45e-53 | 60 | 386 | 112 | 440 | Bacteroidesovatus GH5 xyloglucanase in complex with a XXXG heptasaccharide [Bacteroides ovatus],3ZMR_B Bacteroides ovatus GH5 xyloglucanase in complex with a XXXG heptasaccharide [Bacteroides ovatus] |
| 6XRK_A | 1.58e-51 | 56 | 386 | 17 | 341 | GH5-4broad specificity endoglucanase from an uncultured bovine rumen ciliate [uncultured bovine rumen ciliate],6XRK_B GH5-4 broad specificity endoglucanase from an uncultured bovine rumen ciliate [uncultured bovine rumen ciliate] |
| 2JEP_A | 3.45e-48 | 57 | 386 | 31 | 364 | Nativefamily 5 xyloglucanase from Paenibacillus pabuli [Paenibacillus pabuli],2JEP_B Native family 5 xyloglucanase from Paenibacillus pabuli [Paenibacillus pabuli],2JEQ_A Family 5 xyloglucanase from Paenibacillus pabuli in complex with ligand [Paenibacillus pabuli] |
| 6MQ4_A | 6.46e-48 | 60 | 367 | 9 | 300 | ChainA, cellulase [Acetivibrio cellulolyticus] |
| 6WQY_A | 2.49e-45 | 64 | 386 | 25 | 348 | ChainA, Cellulase [Phocaeicola salanitronis DSM 18170] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A7LXT7 | 4.25e-52 | 60 | 386 | 147 | 475 | Xyloglucan-specific endo-beta-1,4-glucanase BoGH5A OS=Bacteroides ovatus (strain ATCC 8483 / DSM 1896 / JCM 5824 / BCRC 10623 / CCUG 4943 / NCTC 11153) OX=411476 GN=BACOVA_02653 PE=1 SV=1 |
| O08342 | 7.99e-48 | 61 | 386 | 38 | 369 | Endoglucanase A OS=Paenibacillus barcinonensis OX=198119 GN=celA PE=1 SV=1 |
| P28621 | 1.67e-44 | 60 | 386 | 39 | 341 | Endoglucanase B OS=Clostridium cellulovorans (strain ATCC 35296 / DSM 3052 / OCM 3 / 743B) OX=573061 GN=engB PE=3 SV=1 |
| P17901 | 7.95e-43 | 64 | 388 | 48 | 370 | Endoglucanase A OS=Ruminiclostridium cellulolyticum (strain ATCC 35319 / DSM 5812 / JCM 6584 / H10) OX=394503 GN=celCCA PE=1 SV=1 |
| P23661 | 5.06e-41 | 62 | 351 | 70 | 336 | Endoglucanase B OS=Ruminococcus albus OX=1264 GN=celB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
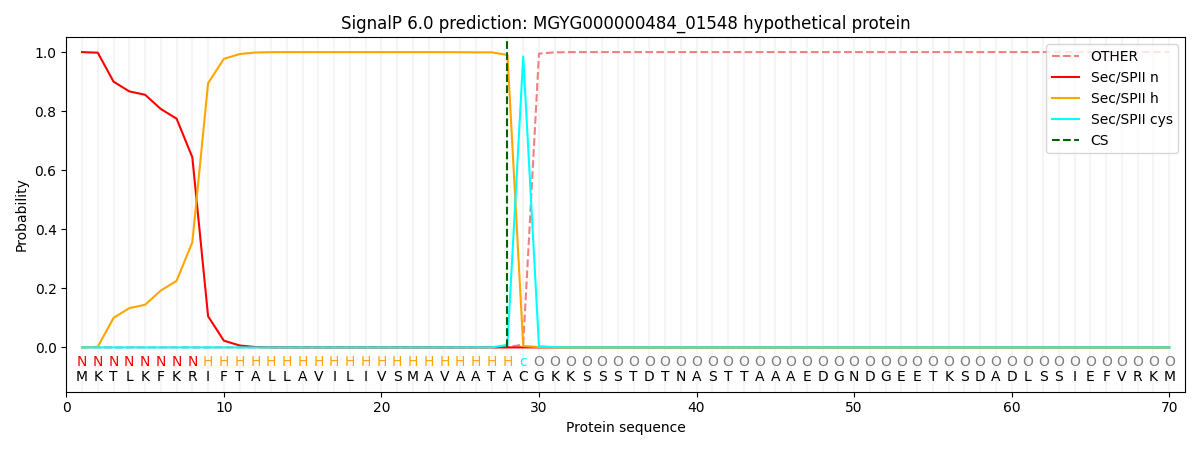
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000001 | 0.000023 | 1.000026 | 0.000000 | 0.000000 | 0.000000 |