You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000497_00602
You are here: Home > Sequence: MGYG000000497_00602
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
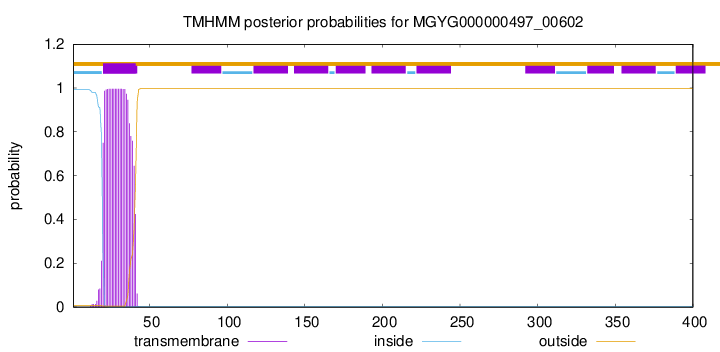
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Opitutales; CAG-312; QALA01; | |||||||||||
| CAZyme ID | MGYG000000497_00602 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | Endoglucanase C307 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 598; End: 1800 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 75 | 367 | 1.7e-87 | 0.9424920127795527 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 3.26e-29 | 79 | 366 | 20 | 268 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 2.46e-13 | 75 | 256 | 66 | 280 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| COG2247 | LytB | 0.008 | 187 | 225 | 49 | 85 | Putative cell wall-binding domain [Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM45789.1 | 2.05e-155 | 41 | 399 | 4 | 364 |
| BAJ62400.1 | 1.37e-74 | 42 | 366 | 5 | 320 |
| CUH93289.1 | 9.90e-74 | 41 | 366 | 4 | 317 |
| AOA60286.1 | 2.03e-73 | 41 | 399 | 4 | 344 |
| AEV59734.1 | 3.04e-70 | 40 | 395 | 4 | 344 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1CEC_A | 4.80e-69 | 44 | 395 | 4 | 338 | ChainA, ENDOGLUCANASE CELC [Acetivibrio thermocellus] |
| 1CEN_A | 1.34e-68 | 44 | 395 | 4 | 338 | ChainA, CELLULASE CELC [Acetivibrio thermocellus],1CEO_A Chain A, CELLULASE CELC [Acetivibrio thermocellus] |
| 3NCO_A | 8.10e-25 | 82 | 366 | 41 | 295 | Crystalstructure of FnCel5A from F. nodosum Rt17-B1 [Fervidobacterium nodosum Rt17-B1],3NCO_B Crystal structure of FnCel5A from F. nodosum Rt17-B1 [Fervidobacterium nodosum Rt17-B1] |
| 3RJY_A | 1.53e-24 | 82 | 366 | 41 | 295 | CrystalStructure of Hyperthermophilic Endo-beta-1,4-glucanase in complex with substrate [Fervidobacterium nodosum Rt17-B1] |
| 3RJX_A | 1.53e-24 | 82 | 366 | 41 | 295 | CrystalStructure of Hyperthermophilic Endo-Beta-1,4-glucanase [Fervidobacterium nodosum Rt17-B1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P23340 | 2.38e-69 | 44 | 395 | 4 | 338 | Endoglucanase C307 OS=Clostridium sp. (strain F1) OX=1508 GN=celC307 PE=1 SV=1 |
| A3DJ77 | 2.38e-69 | 44 | 395 | 4 | 338 | Endoglucanase C OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celC PE=3 SV=1 |
| P0C2S3 | 2.63e-68 | 44 | 395 | 4 | 338 | Endoglucanase C OS=Acetivibrio thermocellus OX=1515 GN=celC PE=1 SV=1 |
| P16169 | 2.23e-39 | 42 | 359 | 5 | 286 | Cellodextrinase A OS=Ruminococcus flavefaciens OX=1265 GN=celA PE=3 SV=3 |
| P25472 | 2.23e-19 | 95 | 259 | 69 | 228 | Endoglucanase D OS=Ruminiclostridium cellulolyticum (strain ATCC 35319 / DSM 5812 / JCM 6584 / H10) OX=394503 GN=celCCD PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
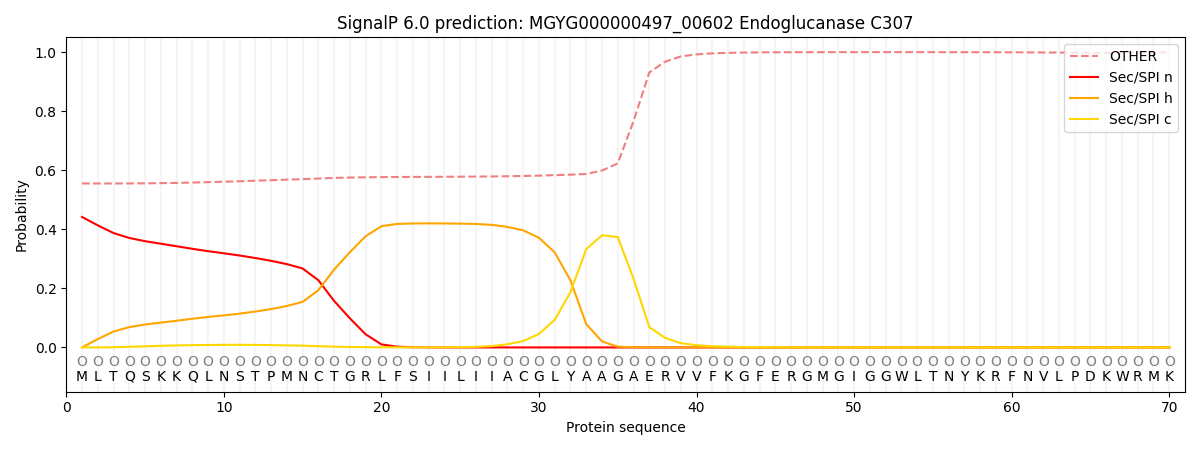
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.573824 | 0.419323 | 0.004108 | 0.000547 | 0.000441 | 0.001756 |