You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000504_00358
You are here: Home > Sequence: MGYG000000504_00358
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-312 sp000438015 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Opitutales; CAG-312; CAG-312; CAG-312 sp000438015 | |||||||||||
| CAZyme ID | MGYG000000504_00358 | |||||||||||
| CAZy Family | GH109 | |||||||||||
| CAZyme Description | Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 124649; End: 125968 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH109 | 31 | 207 | 3.5e-16 | 0.40100250626566414 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0673 | MviM | 2.97e-32 | 32 | 394 | 3 | 337 | Predicted dehydrogenase [General function prediction only]. |
| pfam01408 | GFO_IDH_MocA | 3.38e-13 | 33 | 169 | 1 | 115 | Oxidoreductase family, NAD-binding Rossmann fold. This family of enzymes utilize NADP or NAD. This family is called the GFO/IDH/MOCA family in swiss-prot. |
| TIGR04380 | myo_inos_iolG | 1.99e-10 | 32 | 200 | 1 | 145 | inositol 2-dehydrogenase. All members of the seed alignment for this model are known or predicted inositol 2-dehydrogenase sequences co-clustered with other enzymes for catabolism of myo-inositol or closely related compounds. Inositol 2-dehydrogenase catalyzes the first step in inositol catabolism. Members of this family may vary somewhat in their ranges of acceptable substrates and some may act on analogs to myo-inositol rather than myo-inositol per se. [Energy metabolism, Sugars] |
| PRK11579 | PRK11579 | 9.61e-09 | 30 | 200 | 2 | 145 | putative oxidoreductase; Provisional |
| PRK10206 | PRK10206 | 1.32e-07 | 108 | 201 | 55 | 146 | putative oxidoreductase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AOS46431.1 | 5.76e-104 | 2 | 439 | 10 | 432 |
| AWI08226.1 | 1.08e-103 | 1 | 439 | 4 | 430 |
| AQT66975.1 | 3.70e-103 | 2 | 439 | 12 | 425 |
| AMV39775.1 | 8.37e-98 | 2 | 427 | 5 | 415 |
| VIP04680.1 | 2.00e-97 | 2 | 435 | 10 | 423 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4N54_A | 2.24e-10 | 29 | 329 | 11 | 270 | ChainA, Inositol dehydrogenase [Lacticaseibacillus casei BL23],4N54_B Chain B, Inositol dehydrogenase [Lacticaseibacillus casei BL23],4N54_C Chain C, Inositol dehydrogenase [Lacticaseibacillus casei BL23],4N54_D Chain D, Inositol dehydrogenase [Lacticaseibacillus casei BL23] |
| 4MKX_A | 2.27e-09 | 33 | 329 | 18 | 273 | ChainA, Inositol dehydrogenase [Lacticaseibacillus casei BL23],4MKZ_A Chain A, Inositol dehydrogenase [Lacticaseibacillus casei BL23] |
| 3F4L_A | 3.76e-07 | 108 | 208 | 56 | 154 | Crystalstructure of a probable oxidoreductase yhhX in Triclinic form. Northeast Structural Genomics target ER647 [Escherichia coli K-12],3F4L_B Crystal structure of a probable oxidoreductase yhhX in Triclinic form. Northeast Structural Genomics target ER647 [Escherichia coli K-12],3F4L_C Crystal structure of a probable oxidoreductase yhhX in Triclinic form. Northeast Structural Genomics target ER647 [Escherichia coli K-12],3F4L_D Crystal structure of a probable oxidoreductase yhhX in Triclinic form. Northeast Structural Genomics target ER647 [Escherichia coli K-12],3F4L_E Crystal structure of a probable oxidoreductase yhhX in Triclinic form. Northeast Structural Genomics target ER647 [Escherichia coli K-12],3F4L_F Crystal structure of a probable oxidoreductase yhhX in Triclinic form. Northeast Structural Genomics target ER647 [Escherichia coli K-12] |
| 3GDO_A | 1.23e-06 | 117 | 251 | 66 | 188 | Crystalstructure of putative oxidoreductase yvaA from Bacillus subtilis [Bacillus subtilis subsp. subtilis str. 168],3GDO_B Crystal structure of putative oxidoreductase yvaA from Bacillus subtilis [Bacillus subtilis subsp. subtilis str. 168] |
| 3GFG_A | 1.26e-06 | 117 | 251 | 75 | 197 | Structureof putative oxidoreductase yvaA from Bacillus subtilis in triclinic form [Bacillus subtilis subsp. subtilis str. 168],3GFG_B Structure of putative oxidoreductase yvaA from Bacillus subtilis in triclinic form [Bacillus subtilis subsp. subtilis str. 168],3GFG_C Structure of putative oxidoreductase yvaA from Bacillus subtilis in triclinic form [Bacillus subtilis subsp. subtilis str. 168],3GFG_D Structure of putative oxidoreductase yvaA from Bacillus subtilis in triclinic form [Bacillus subtilis subsp. subtilis str. 168],3GFG_E Structure of putative oxidoreductase yvaA from Bacillus subtilis in triclinic form [Bacillus subtilis subsp. subtilis str. 168],3GFG_F Structure of putative oxidoreductase yvaA from Bacillus subtilis in triclinic form [Bacillus subtilis subsp. subtilis str. 168],3GFG_G Structure of putative oxidoreductase yvaA from Bacillus subtilis in triclinic form [Bacillus subtilis subsp. subtilis str. 168],3GFG_H Structure of putative oxidoreductase yvaA from Bacillus subtilis in triclinic form [Bacillus subtilis subsp. subtilis str. 168],3GFG_I Structure of putative oxidoreductase yvaA from Bacillus subtilis in triclinic form [Bacillus subtilis subsp. subtilis str. 168],3GFG_J Structure of putative oxidoreductase yvaA from Bacillus subtilis in triclinic form [Bacillus subtilis subsp. subtilis str. 168],3GFG_K Structure of putative oxidoreductase yvaA from Bacillus subtilis in triclinic form [Bacillus subtilis subsp. subtilis str. 168],3GFG_L Structure of putative oxidoreductase yvaA from Bacillus subtilis in triclinic form [Bacillus subtilis subsp. subtilis str. 168] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P74041 | 3.06e-07 | 106 | 348 | 57 | 276 | Uncharacterized oxidoreductase sll0816 OS=Synechocystis sp. (strain PCC 6803 / Kazusa) OX=1111708 GN=sll0816 PE=3 SV=1 |
| B5Y2S5 | 4.78e-07 | 107 | 269 | 57 | 194 | Inositol 2-dehydrogenase OS=Klebsiella pneumoniae (strain 342) OX=507522 GN=iolG PE=3 SV=1 |
| A0KYQ9 | 4.97e-07 | 2 | 205 | 5 | 210 | Glycosyl hydrolase family 109 protein 2 OS=Shewanella sp. (strain ANA-3) OX=94122 GN=Shewana3_2701 PE=3 SV=1 |
| Q0HH61 | 4.97e-07 | 2 | 205 | 5 | 210 | Glycosyl hydrolase family 109 protein 2 OS=Shewanella sp. (strain MR-4) OX=60480 GN=Shewmr4_2535 PE=3 SV=1 |
| Q0HTG8 | 4.97e-07 | 2 | 205 | 5 | 210 | Glycosyl hydrolase family 109 protein 2 OS=Shewanella sp. (strain MR-7) OX=60481 GN=Shewmr7_2602 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as TAT
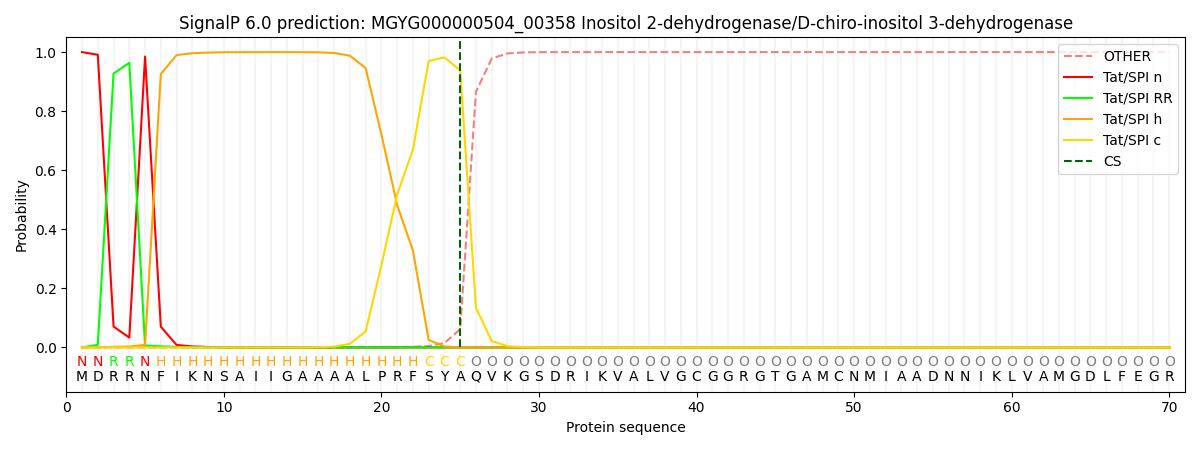
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 0.000000 | 0.999972 | 0.000000 | 0.000000 |
