You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000504_00998
You are here: Home > Sequence: MGYG000000504_00998
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-312 sp000438015 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Opitutales; CAG-312; CAG-312; CAG-312 sp000438015 | |||||||||||
| CAZyme ID | MGYG000000504_00998 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 125890; End: 127329 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 66 | 323 | 5.9e-22 | 0.6363636363636364 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBL08912.1 | 4.02e-116 | 26 | 478 | 32 | 469 |
| BBL11704.1 | 4.02e-116 | 26 | 478 | 32 | 469 |
| BBL00987.1 | 1.14e-115 | 26 | 478 | 32 | 469 |
| AEI51976.1 | 1.75e-110 | 26 | 477 | 29 | 474 |
| AXE20299.1 | 3.93e-109 | 26 | 477 | 29 | 474 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3ZIZ_A | 4.89e-07 | 70 | 235 | 49 | 212 | ChainA, Gh5 Endo-beta-1,4-mannanase [Podospora anserina] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9FJZ3 | 6.06e-09 | 50 | 348 | 47 | 362 | Mannan endo-1,4-beta-mannosidase 7 OS=Arabidopsis thaliana OX=3702 GN=MAN7 PE=2 SV=1 |
| Q10B67 | 8.63e-09 | 70 | 269 | 62 | 276 | Mannan endo-1,4-beta-mannosidase 4 OS=Oryza sativa subsp. japonica OX=39947 GN=MAN4 PE=2 SV=2 |
| A1D8Y6 | 4.89e-08 | 1 | 234 | 1 | 223 | Probable mannan endo-1,4-beta-mannosidase A OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=manA PE=3 SV=1 |
| Q0CUG5 | 5.29e-08 | 49 | 245 | 62 | 261 | Probable mannan endo-1,4-beta-mannosidase A-2 OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=manA-2 PE=3 SV=2 |
| Q5AVP1 | 4.88e-07 | 70 | 235 | 61 | 224 | Mannan endo-1,4-beta-mannosidase D OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=manD PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
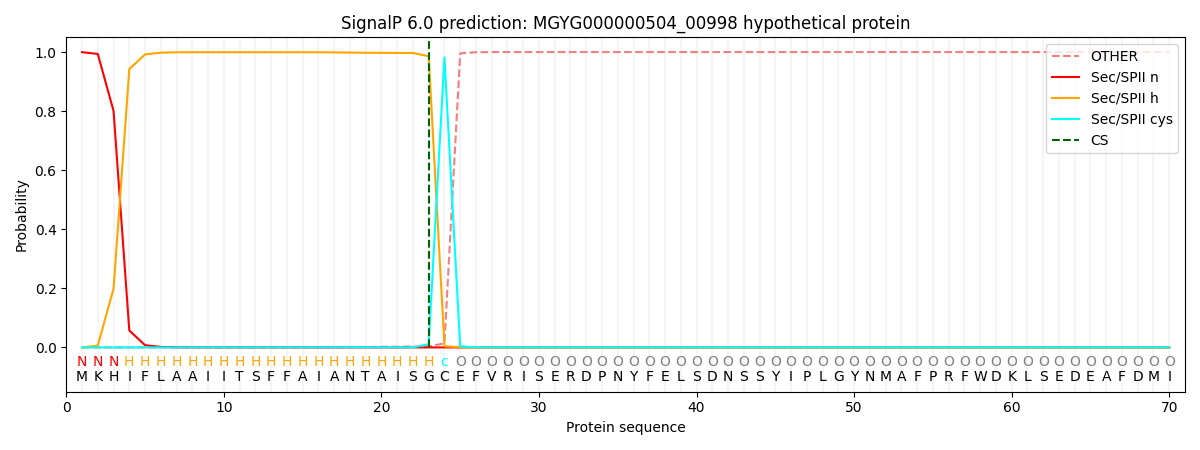
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000160 | 0.999872 | 0.000000 | 0.000000 | 0.000000 |
