You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000504_01154
You are here: Home > Sequence: MGYG000000504_01154
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-312 sp000438015 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Verrucomicrobiae; Opitutales; CAG-312; CAG-312; CAG-312 sp000438015 | |||||||||||
| CAZyme ID | MGYG000000504_01154 | |||||||||||
| CAZy Family | GH110 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 51756; End: 54128 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH110 | 215 | 757 | 5.7e-49 | 0.9945255474452555 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13229 | Beta_helix | 5.76e-09 | 588 | 758 | 28 | 157 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam13229 | Beta_helix | 1.88e-07 | 589 | 758 | 6 | 133 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| COG5434 | Pgu1 | 3.43e-05 | 579 | 757 | 287 | 451 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| TIGR03804 | para_beta_helix | 0.001 | 714 | 758 | 3 | 38 | parallel beta-helix repeat (two copies). This model represents a tandem pair of an approximately 22-amino acid (each) repeat homologous to the beta-strand repeats that stack in a right-handed parallel beta-helix in the periplasmic C-5 mannuronan epimerase, AlgA, of Pseudomonas aeruginosa. A homology domain consisting of a longer tandem array of these repeats is described in the SMART database as CASH (SM00722), and is found in many carbohydrate-binding proteins and sugar hydrolases. A single repeat is represented by SM00710. This TIGRFAMs model represents a flavor of the parallel beta-helix-forming repeat based on prokaryotic sequences only in its seed alignment, although it also finds many eukaryotic sequences. |
| pfam13229 | Beta_helix | 0.001 | 696 | 759 | 9 | 64 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM44429.1 | 1.59e-196 | 49 | 790 | 42 | 770 |
| AVM44109.1 | 2.88e-192 | 49 | 790 | 83 | 807 |
| AVM44539.1 | 3.34e-131 | 50 | 789 | 44 | 764 |
| AVM46987.1 | 2.61e-105 | 57 | 789 | 20 | 746 |
| AZM48752.1 | 6.74e-87 | 193 | 789 | 39 | 621 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7JW4_A | 1.15e-12 | 208 | 729 | 22 | 598 | Crystalstructure of PdGH110B in complex with D-galactose [Pseudoalteromonas distincta],7JW4_B Crystal structure of PdGH110B in complex with D-galactose [Pseudoalteromonas distincta] |
| 7JWF_A | 4.59e-12 | 208 | 729 | 22 | 598 | Crystalstructure of PdGH110B D344N in complex with alpha-(1,3)-galactobiose [Pseudoalteromonas distincta],7JWF_B Crystal structure of PdGH110B D344N in complex with alpha-(1,3)-galactobiose [Pseudoalteromonas distincta],7JWF_C Crystal structure of PdGH110B D344N in complex with alpha-(1,3)-galactobiose [Pseudoalteromonas distincta],7JWF_D Crystal structure of PdGH110B D344N in complex with alpha-(1,3)-galactobiose [Pseudoalteromonas distincta] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q5L7M8 | 1.62e-16 | 208 | 753 | 20 | 572 | Alpha-1,3-galactosidase A OS=Bacteroides fragilis (strain ATCC 25285 / DSM 2151 / CCUG 4856 / JCM 11019 / NCTC 9343 / Onslow) OX=272559 GN=glaA PE=3 SV=1 |
| Q64MU6 | 2.83e-16 | 208 | 753 | 20 | 572 | Alpha-1,3-galactosidase A OS=Bacteroides fragilis (strain YCH46) OX=295405 GN=glaA PE=3 SV=1 |
| B1KD83 | 2.24e-13 | 214 | 760 | 33 | 583 | Alpha-1,3-galactosidase A OS=Shewanella woodyi (strain ATCC 51908 / MS32) OX=392500 GN=glaA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
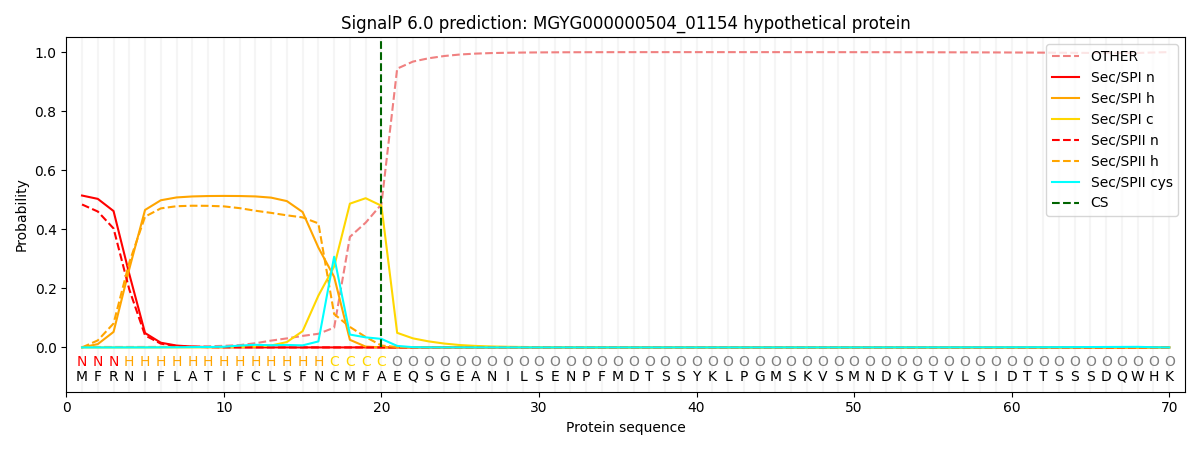
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002888 | 0.502976 | 0.493178 | 0.000463 | 0.000253 | 0.000228 |
