You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000510_00905
You are here: Home > Sequence: MGYG000000510_00905
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UMGS363 sp900541495 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; UMGS363; UMGS363 sp900541495 | |||||||||||
| CAZyme ID | MGYG000000510_00905 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 42354; End: 43514 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 61 | 350 | 4.1e-44 | 0.980327868852459 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBL16471.1 | 4.57e-113 | 45 | 386 | 29 | 363 |
| ADD61974.1 | 4.64e-111 | 43 | 386 | 65 | 407 |
| CCO03823.1 | 5.62e-110 | 43 | 386 | 56 | 398 |
| ADU21807.1 | 3.66e-102 | 46 | 386 | 72 | 409 |
| AGS51602.1 | 3.78e-96 | 16 | 349 | 13 | 357 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4OOU_A | 6.24e-11 | 53 | 368 | 27 | 362 | Crystalstructure of beta-1,4-D-mannanase from Cryptopygus antarcticus [Cryptopygus antarcticus],4OOU_B Crystal structure of beta-1,4-D-mannanase from Cryptopygus antarcticus [Cryptopygus antarcticus],4OOZ_A Crystal structure of beta-1,4-D-mannanase from Cryptopygus antarcticus in complex with mannopentaose [Cryptopygus antarcticus],4OOZ_B Crystal structure of beta-1,4-D-mannanase from Cryptopygus antarcticus in complex with mannopentaose [Cryptopygus antarcticus] |
| 1RH9_A | 7.46e-06 | 49 | 318 | 7 | 292 | ChainA, endo-beta-mannanase [Solanum lycopersicum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B4XC07 | 3.34e-10 | 53 | 368 | 27 | 362 | Mannan endo-1,4-beta-mannosidase OS=Cryptopygus antarcticus OX=187623 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
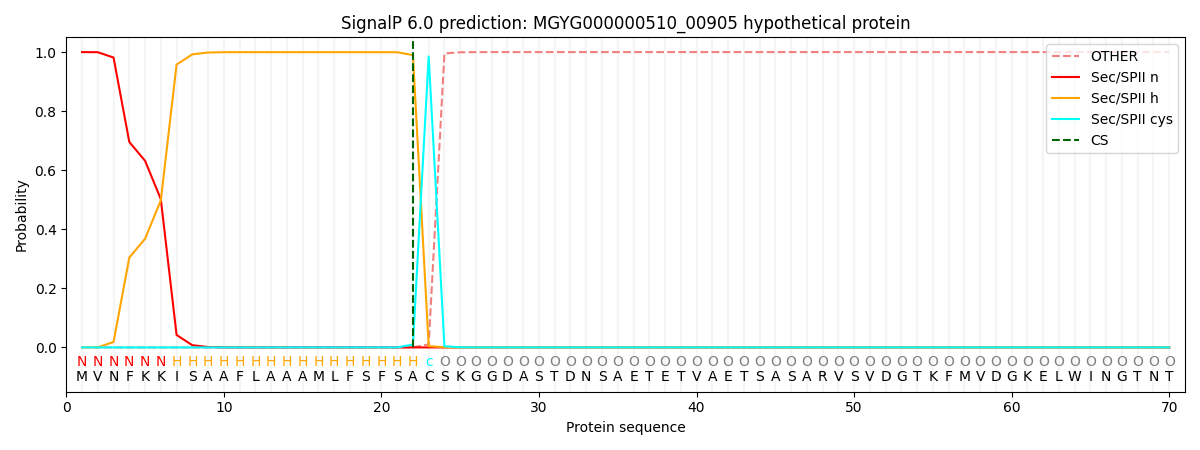
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000123 | 0.999935 | 0.000000 | 0.000000 | 0.000000 |
