You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000524_01280
You are here: Home > Sequence: MGYG000000524_01280
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
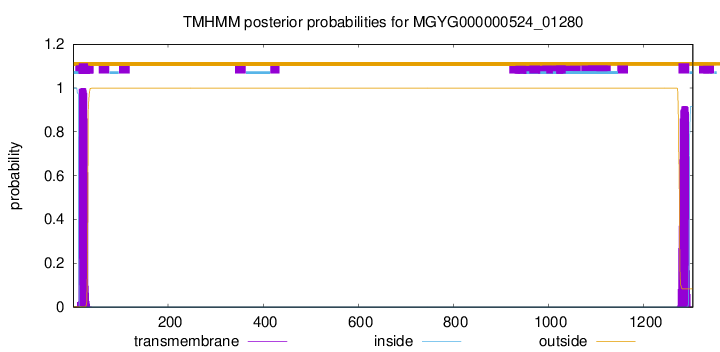
TMHMM annotations
Basic Information help
| Species | UBA4636 sp900770945 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia_A; Christensenellales; Borkfalkiaceae; UBA4636; UBA4636 sp900770945 | |||||||||||
| CAZyme ID | MGYG000000524_01280 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 4692; End: 8606 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK15098 | PRK15098 | 7.09e-18 | 380 | 529 | 631 | 764 | beta-glucosidase BglX. |
| NF033189 | internalin_A | 6.41e-12 | 998 | 1266 | 445 | 707 | class 1 internalin InlA. Internalins, as found in the intracellular human pathogen Listeria monocytogenes, are paralogous surface or secreted proteins with an N-terminal signal peptide, leucine-rich repeats, and usually a C-terminal LPXTG processing and cell surface anchoring site. See PMID:17764999 for a general discussion of internalins. Members of this family are internalin A (InlA), a class 1 (LPXTG-type) internalin. |
| pfam09479 | Flg_new | 1.27e-09 | 1132 | 1186 | 12 | 65 | Listeria-Bacteroides repeat domain (List_Bact_rpt). This model describes a conserved core region of about 43 residues, which occurs in at least two families of tandem repeats. These include 78-residue repeats which occur from 2 to 15 times in some proteins of Bacteroides forsythus ATCC 43037, and 70-residue repeats found in families of internalins of Listeria species. Single copies are found in proteins of Fibrobacter succinogenes, Geobacter sulfurreducens, and a few other bacteria. |
| NF033188 | internalin_H | 1.40e-08 | 1113 | 1250 | 340 | 467 | InlH/InlC2 family class 1 internalin. Internalins, as found in the intracellular human pathogen Listeria monocytogenes, are paralogous surface or secreted proteins with an N-terminal signal peptide, leucine-rich repeats, and usually a C-terminal LPXTG processing and cell surface anchoring site. See PMID:17764999 for a general discussion of internalins. Members of this family are internalin H (InlH), or internalin C2, two class 1 (LPXTG-type) internalins that are closely related, one apparently derived from the other through a recombination event. |
| pfam00933 | Glyco_hydro_3 | 2.30e-08 | 751 | 911 | 95 | 257 | Glycosyl hydrolase family 3 N terminal domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QOS39239.1 | 1.00e-132 | 56 | 912 | 65 | 870 |
| VEU80232.1 | 6.56e-132 | 48 | 908 | 70 | 857 |
| VEU80230.1 | 2.09e-128 | 47 | 917 | 28 | 846 |
| QOS39237.1 | 3.61e-121 | 47 | 982 | 46 | 934 |
| AYV69401.1 | 1.62e-113 | 57 | 913 | 56 | 852 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5WUG_A | 4.04e-52 | 62 | 908 | 35 | 747 | Expression,characterization and crystal structure of a novel beta-glucosidase from Paenibacillus barengoltzii [Paenibacillus barengoltzii],5WVP_A Expression, characterization and crystal structure of a novel beta-glucosidase from Paenibacillus barengoltzii [Paenibacillus barengoltzii] |
| 2X40_A | 1.76e-21 | 741 | 930 | 78 | 272 | Structureof beta-glucosidase 3B from Thermotoga neapolitana in complex with glycerol [Thermotoga neapolitana DSM 4359],2X41_A Structure of beta-glucosidase 3B from Thermotoga neapolitana in complex with glucose [Thermotoga neapolitana DSM 4359] |
| 2X42_A | 1.60e-20 | 741 | 930 | 78 | 272 | Structureof beta-glucosidase 3B from Thermotoga neapolitana in complex with alpha-D-glucose [Thermotoga neapolitana DSM 4359] |
| 3AC0_A | 7.95e-18 | 713 | 879 | 28 | 196 | Crystalstructure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus],3AC0_B Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus],3AC0_C Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus],3AC0_D Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus] |
| 3ABZ_A | 1.22e-16 | 713 | 879 | 28 | 196 | Crystalstructure of Se-Met labeled Beta-glucosidase from Kluyveromyces marxianus [Kluyveromyces marxianus],3ABZ_B Crystal structure of Se-Met labeled Beta-glucosidase from Kluyveromyces marxianus [Kluyveromyces marxianus],3ABZ_C Crystal structure of Se-Met labeled Beta-glucosidase from Kluyveromyces marxianus [Kluyveromyces marxianus],3ABZ_D Crystal structure of Se-Met labeled Beta-glucosidase from Kluyveromyces marxianus [Kluyveromyces marxianus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P16084 | 4.61e-52 | 65 | 908 | 31 | 769 | Beta-glucosidase A OS=Butyrivibrio fibrisolvens OX=831 GN=bglA PE=3 SV=1 |
| P15885 | 8.33e-50 | 69 | 908 | 16 | 696 | Beta-glucosidase OS=Ruminococcus albus OX=1264 PE=3 SV=1 |
| Q9P6J6 | 1.04e-24 | 694 | 908 | 8 | 225 | Putative beta-glucosidase OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPBC1683.04 PE=3 SV=1 |
| A1DNN8 | 6.72e-23 | 696 | 908 | 18 | 233 | Probable beta-glucosidase J OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=bglJ PE=3 SV=1 |
| Q4WLY1 | 1.16e-22 | 696 | 908 | 18 | 233 | Probable beta-glucosidase J OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=bglJ PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
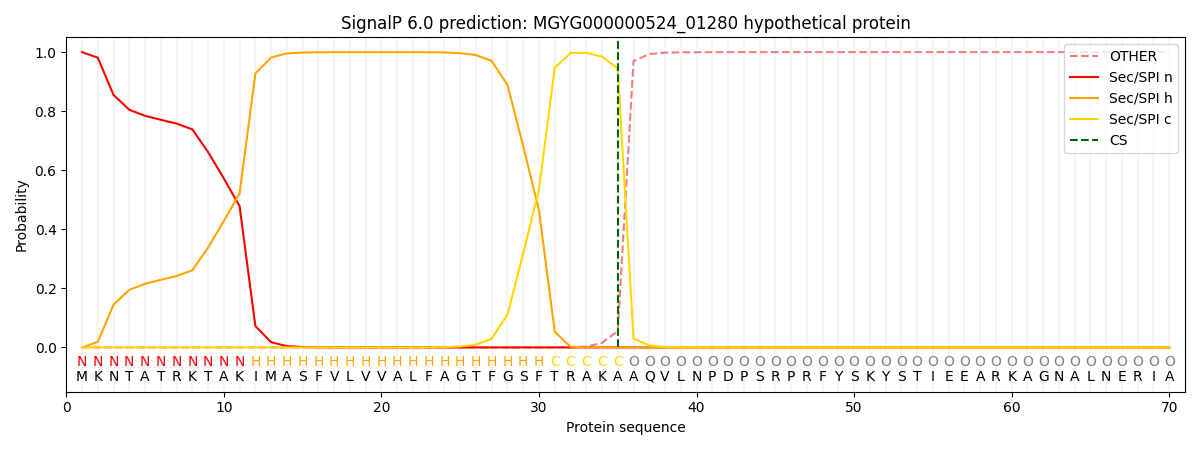
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000578 | 0.998156 | 0.000650 | 0.000223 | 0.000199 | 0.000165 |