You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000526_00588
You are here: Home > Sequence: MGYG000000526_00588
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
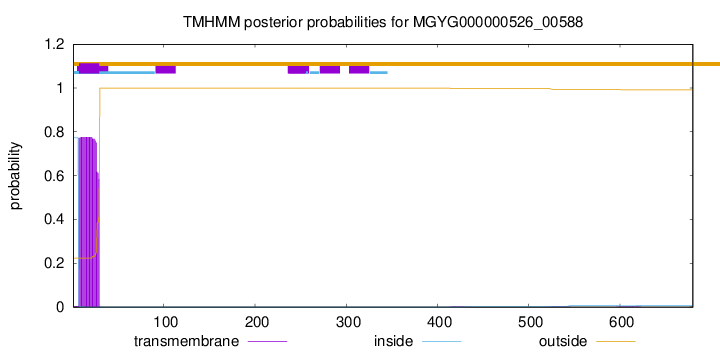
TMHMM annotations
Basic Information help
| Species | Prevotella sp900770395 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900770395 | |||||||||||
| CAZyme ID | MGYG000000526_00588 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 109879; End: 111921 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 49 | 315 | 7.4e-104 | 0.9884169884169884 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 3.07e-14 | 89 | 265 | 32 | 201 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 5.65e-04 | 93 | 246 | 85 | 242 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCD43724.1 | 1.08e-238 | 21 | 680 | 6 | 669 |
| AHW58816.1 | 5.39e-229 | 25 | 679 | 22 | 677 |
| QDO69414.1 | 4.03e-204 | 30 | 611 | 52 | 644 |
| QRX63289.1 | 2.14e-203 | 31 | 610 | 52 | 638 |
| EDV05070.1 | 2.98e-202 | 30 | 611 | 56 | 648 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6GJF_A | 3.29e-09 | 34 | 264 | 13 | 207 | Ancestralendocellulase Cel5A [synthetic construct],6GJF_B Ancestral endocellulase Cel5A [synthetic construct],6GJF_C Ancestral endocellulase Cel5A [synthetic construct],6GJF_D Ancestral endocellulase Cel5A [synthetic construct],6GJF_E Ancestral endocellulase Cel5A [synthetic construct],6GJF_F Ancestral endocellulase Cel5A [synthetic construct] |
| 5IHS_A | 1.03e-08 | 30 | 271 | 31 | 234 | Structureof CHU_2103 from Cytophaga hutchinsonii [Cytophaga hutchinsonii ATCC 33406] |
| 1EGZ_A | 1.30e-06 | 188 | 276 | 123 | 213 | ChainA, ENDOGLUCANASE Z [Dickeya chrysanthemi],1EGZ_B Chain B, ENDOGLUCANASE Z [Dickeya chrysanthemi],1EGZ_C Chain C, ENDOGLUCANASE Z [Dickeya chrysanthemi] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q47096 | 1.83e-09 | 34 | 264 | 41 | 235 | Endoglucanase 5 OS=Pectobacterium carotovorum subsp. carotovorum OX=555 GN=celV PE=1 SV=1 |
| Q59394 | 2.77e-09 | 34 | 264 | 41 | 235 | Endoglucanase N OS=Pectobacterium atrosepticum OX=29471 GN=celN PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
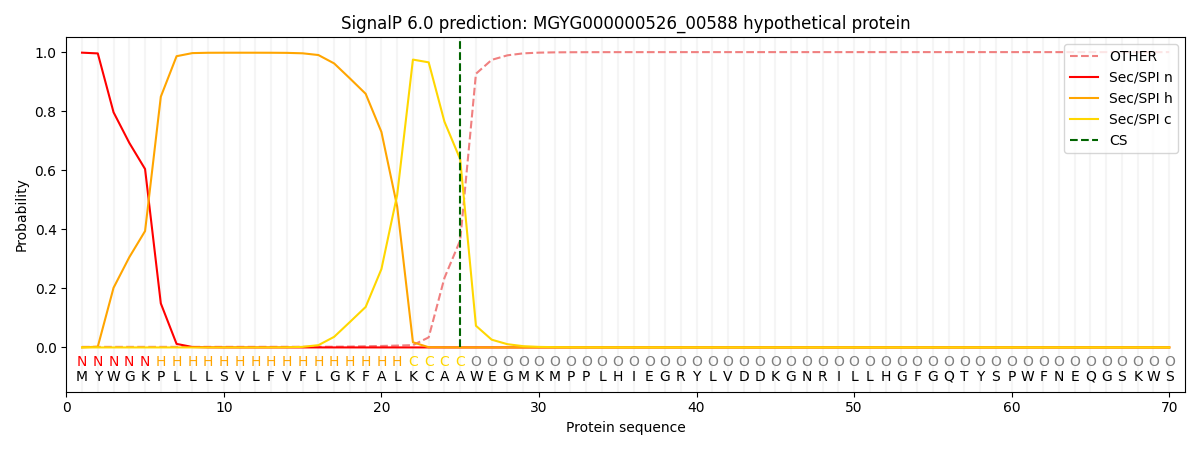
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.003271 | 0.995697 | 0.000409 | 0.000194 | 0.000188 | 0.000208 |