You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000526_01174
You are here: Home > Sequence: MGYG000000526_01174
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp900770395 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900770395 | |||||||||||
| CAZyme ID | MGYG000000526_01174 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 150801; End: 152189 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 70 | 377 | 1.5e-88 | 0.9855072463768116 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 6.97e-47 | 70 | 376 | 15 | 266 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 4.89e-19 | 34 | 376 | 48 | 357 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADE83418.1 | 3.29e-150 | 26 | 405 | 24 | 394 |
| AAC36862.1 | 3.29e-150 | 26 | 405 | 24 | 394 |
| QVJ80006.1 | 5.14e-149 | 1 | 405 | 1 | 393 |
| AIF25923.1 | 3.42e-147 | 26 | 405 | 10 | 391 |
| ACA61149.1 | 5.53e-141 | 27 | 403 | 144 | 519 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6XSO_A | 1.29e-139 | 27 | 405 | 19 | 378 | GH5-4broad specificity endoglucanase from an uncultured bacterium [uncultured bacterium],6XSO_B GH5-4 broad specificity endoglucanase from an uncultured bacterium [uncultured bacterium] |
| 5D9N_A | 6.46e-80 | 28 | 403 | 11 | 351 | Crystalstructure of PbGH5A, a glycoside hydrolase family 5 member from Prevotella bryantii B14, in complex with the xyloglucan heptasaccharide XXXG [Prevotella bryantii],5D9N_B Crystal structure of PbGH5A, a glycoside hydrolase family 5 member from Prevotella bryantii B14, in complex with the xyloglucan heptasaccharide XXXG [Prevotella bryantii],5D9P_A Crystal structure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14, in complex with an inhibitory N-bromoacetylglycosylamine derivative of XXXG [Prevotella bryantii],5D9P_B Crystal structure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14, in complex with an inhibitory N-bromoacetylglycosylamine derivative of XXXG [Prevotella bryantii] |
| 5D9M_A | 5.08e-79 | 28 | 403 | 11 | 351 | Crystalstructure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14, E280A mutant in complex with the xyloglucan tetradecasaccharide XXXGXXXG [Prevotella bryantii],5D9M_B Crystal structure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14, E280A mutant in complex with the xyloglucan tetradecasaccharide XXXGXXXG [Prevotella bryantii],5D9O_A Crystal structure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14, E280A mutant in complex with cellotetraose [Prevotella bryantii],5D9O_B Crystal structure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14, E280A mutant in complex with cellotetraose [Prevotella bryantii] |
| 6D2W_A | 1.35e-75 | 28 | 403 | 434 | 774 | Crystalstructure of Prevotella bryantii endo-beta-mannanase/endo-beta-glucanase PbGH26A-GH5A [Prevotella bryantii B14],6D2W_B Crystal structure of Prevotella bryantii endo-beta-mannanase/endo-beta-glucanase PbGH26A-GH5A [Prevotella bryantii B14] |
| 3VDH_A | 2.70e-75 | 28 | 403 | 11 | 351 | Crystalstructure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14 [Prevotella bryantii],3VDH_B Crystal structure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14 [Prevotella bryantii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q12647 | 1.29e-46 | 29 | 402 | 24 | 362 | Endoglucanase B OS=Neocallimastix patriciarum OX=4758 GN=CELB PE=2 SV=1 |
| P10477 | 6.46e-43 | 29 | 376 | 56 | 349 | Cellulase/esterase CelE OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celE PE=1 SV=2 |
| P54937 | 2.29e-40 | 29 | 403 | 38 | 378 | Endoglucanase A OS=Clostridium longisporum OX=1523 GN=celA PE=1 SV=1 |
| P23660 | 1.01e-39 | 29 | 364 | 26 | 318 | Endoglucanase A OS=Ruminococcus albus OX=1264 GN=celA PE=1 SV=1 |
| P17901 | 4.19e-39 | 34 | 441 | 51 | 440 | Endoglucanase A OS=Ruminiclostridium cellulolyticum (strain ATCC 35319 / DSM 5812 / JCM 6584 / H10) OX=394503 GN=celCCA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
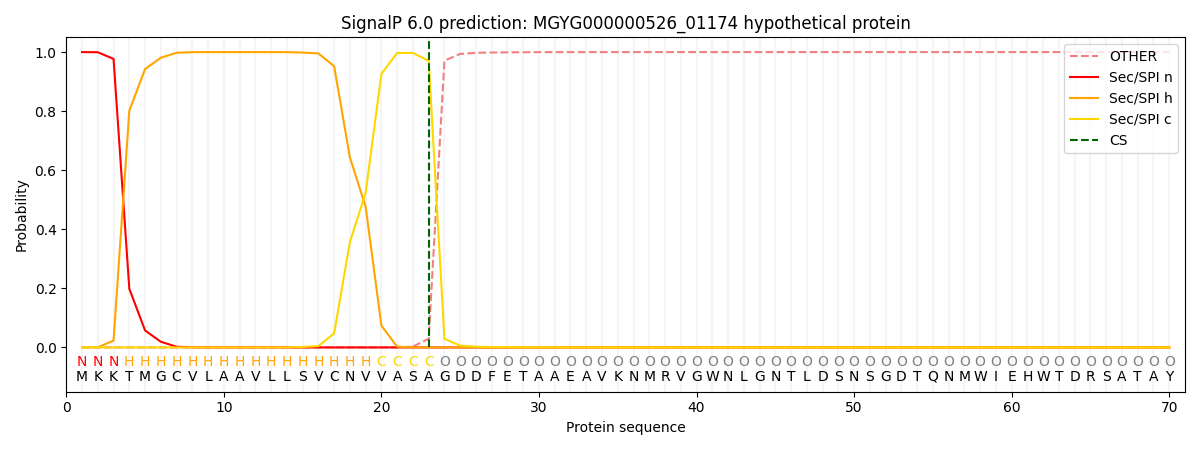
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000283 | 0.998930 | 0.000323 | 0.000167 | 0.000154 | 0.000149 |
