You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000526_02745
You are here: Home > Sequence: MGYG000000526_02745
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp900770395 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900770395 | |||||||||||
| CAZyme ID | MGYG000000526_02745 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | Endoglucanase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 405; End: 1367 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 42 | 275 | 1.6e-87 | 0.9873417721518988 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 3.20e-51 | 42 | 282 | 4 | 271 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 6.11e-05 | 19 | 209 | 35 | 250 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDH78944.1 | 5.44e-110 | 23 | 316 | 28 | 319 |
| ALJ01521.1 | 6.43e-110 | 3 | 315 | 4 | 316 |
| ADD61407.1 | 9.07e-110 | 31 | 313 | 22 | 303 |
| ADD62012.1 | 9.07e-110 | 31 | 313 | 22 | 303 |
| BBK85897.1 | 1.31e-109 | 31 | 318 | 30 | 316 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5IHS_A | 7.98e-74 | 6 | 313 | 6 | 317 | Structureof CHU_2103 from Cytophaga hutchinsonii [Cytophaga hutchinsonii ATCC 33406] |
| 6GJF_A | 2.20e-68 | 23 | 313 | 5 | 297 | Ancestralendocellulase Cel5A [synthetic construct],6GJF_B Ancestral endocellulase Cel5A [synthetic construct],6GJF_C Ancestral endocellulase Cel5A [synthetic construct],6GJF_D Ancestral endocellulase Cel5A [synthetic construct],6GJF_E Ancestral endocellulase Cel5A [synthetic construct],6GJF_F Ancestral endocellulase Cel5A [synthetic construct] |
| 3PZT_A | 1.99e-66 | 22 | 313 | 28 | 321 | Structureof the endo-1,4-beta-glucanase from Bacillus subtilis 168 with manganese(II) ion [Bacillus subtilis subsp. subtilis str. 168],3PZT_B Structure of the endo-1,4-beta-glucanase from Bacillus subtilis 168 with manganese(II) ion [Bacillus subtilis subsp. subtilis str. 168],3PZU_A P212121 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZU_B P212121 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_A C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_B C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_C C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_D C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168] |
| 4XZW_A | 7.44e-66 | 20 | 305 | 1 | 291 | Endo-glucanasechimera C10 [uncultured bacterium] |
| 5WH8_A | 5.58e-65 | 23 | 300 | 6 | 293 | CellulaseCel5C_n [uncultured organism] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P06565 | 8.01e-64 | 5 | 313 | 12 | 323 | Endoglucanase B OS=Evansella cellulosilytica (strain ATCC 21833 / DSM 2522 / FERM P-1141 / JCM 9156 / N-4) OX=649639 GN=celB PE=3 SV=1 |
| P10475 | 9.02e-64 | 22 | 313 | 33 | 326 | Endoglucanase OS=Bacillus subtilis (strain 168) OX=224308 GN=eglS PE=1 SV=1 |
| P07983 | 6.92e-63 | 22 | 297 | 33 | 311 | Endoglucanase OS=Bacillus subtilis OX=1423 GN=bglC PE=3 SV=2 |
| O85465 | 7.68e-62 | 5 | 313 | 12 | 323 | Endoglucanase 5A OS=Salipaludibacillus agaradhaerens OX=76935 GN=cel5A PE=1 SV=1 |
| P23549 | 1.04e-61 | 5 | 313 | 16 | 326 | Endoglucanase OS=Bacillus subtilis OX=1423 GN=bglC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
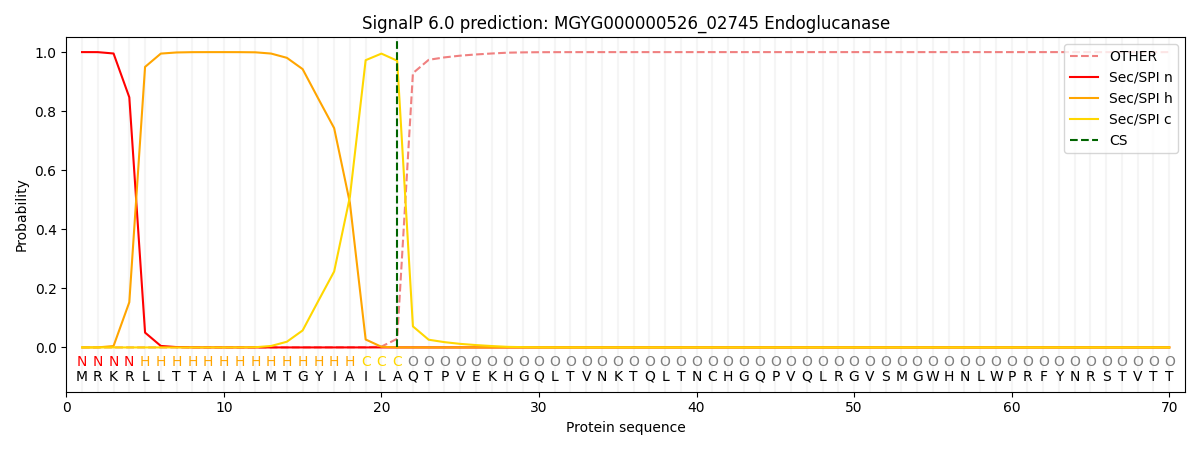
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000700 | 0.998527 | 0.000247 | 0.000181 | 0.000168 | 0.000155 |
