You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000532_01609
You are here: Home > Sequence: MGYG000000532_01609
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
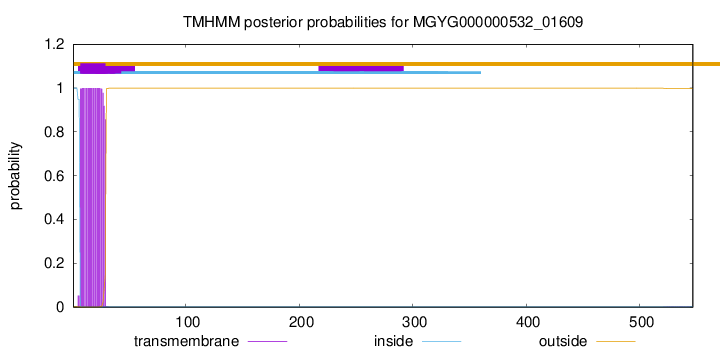
TMHMM annotations
Basic Information help
| Species | CAG-510 sp000434615 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; CAG-510; CAG-510 sp000434615 | |||||||||||
| CAZyme ID | MGYG000000532_01609 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 55781; End: 57424 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 123 | 364 | 4.6e-96 | 0.9915611814345991 |
| CBM2 | 455 | 540 | 3.8e-17 | 0.7920792079207921 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 3.99e-67 | 121 | 372 | 1 | 272 | Cellulase (glycosyl hydrolase family 5). |
| smart00637 | CBD_II | 4.91e-13 | 459 | 547 | 2 | 83 | CBD_II domain. |
| pfam00553 | CBM_2 | 3.30e-11 | 454 | 544 | 4 | 89 | Cellulose binding domain. Two tryptophan residues are involved in cellulose binding. Cellulose binding domain found in bacteria. |
| COG2730 | BglC | 2.83e-06 | 113 | 414 | 47 | 407 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| pfam09478 | CBM49 | 1.48e-04 | 459 | 538 | 9 | 80 | Carbohydrate binding domain CBM49. This domain is found at the C terminal of cellulases and in vitro binding studies have shown it to binds to crystalline cellulose. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AFA47670.1 | 7.63e-141 | 111 | 540 | 203 | 626 |
| SIP56455.1 | 5.46e-140 | 104 | 409 | 78 | 382 |
| SIP56505.1 | 1.09e-139 | 104 | 409 | 78 | 382 |
| SIP56560.1 | 1.09e-139 | 104 | 409 | 78 | 382 |
| CBK89462.1 | 2.52e-139 | 104 | 409 | 82 | 386 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6GJF_A | 1.23e-97 | 111 | 404 | 11 | 300 | Ancestralendocellulase Cel5A [synthetic construct],6GJF_B Ancestral endocellulase Cel5A [synthetic construct],6GJF_C Ancestral endocellulase Cel5A [synthetic construct],6GJF_D Ancestral endocellulase Cel5A [synthetic construct],6GJF_E Ancestral endocellulase Cel5A [synthetic construct],6GJF_F Ancestral endocellulase Cel5A [synthetic construct] |
| 4XZB_A | 9.25e-88 | 111 | 404 | 10 | 303 | endo-glucanaseGsCelA P1 [Geobacillus sp. 70PC53] |
| 4XZW_A | 3.92e-86 | 111 | 404 | 10 | 302 | Endo-glucanasechimera C10 [uncultured bacterium] |
| 3PZT_A | 1.89e-84 | 111 | 406 | 35 | 326 | Structureof the endo-1,4-beta-glucanase from Bacillus subtilis 168 with manganese(II) ion [Bacillus subtilis subsp. subtilis str. 168],3PZT_B Structure of the endo-1,4-beta-glucanase from Bacillus subtilis 168 with manganese(II) ion [Bacillus subtilis subsp. subtilis str. 168],3PZU_A P212121 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZU_B P212121 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_A C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_B C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_C C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_D C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168] |
| 5WH8_A | 2.84e-82 | 111 | 404 | 12 | 307 | CellulaseCel5C_n [uncultured organism] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P22541 | 1.24e-102 | 111 | 400 | 114 | 403 | Endoglucanase A OS=Butyrivibrio fibrisolvens OX=831 GN=celA PE=1 SV=1 |
| Q07940 | 4.03e-95 | 127 | 398 | 18 | 288 | Endoglucanase 4 OS=Ruminococcus albus OX=1264 GN=Eg IV PE=1 SV=1 |
| P15704 | 1.71e-91 | 103 | 410 | 37 | 341 | Endoglucanase OS=Clostridium saccharobutylicum OX=169679 GN=eglA PE=3 SV=1 |
| P10475 | 7.75e-82 | 111 | 415 | 40 | 338 | Endoglucanase OS=Bacillus subtilis (strain 168) OX=224308 GN=eglS PE=1 SV=1 |
| P07983 | 2.15e-81 | 111 | 415 | 40 | 338 | Endoglucanase OS=Bacillus subtilis OX=1423 GN=bglC PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
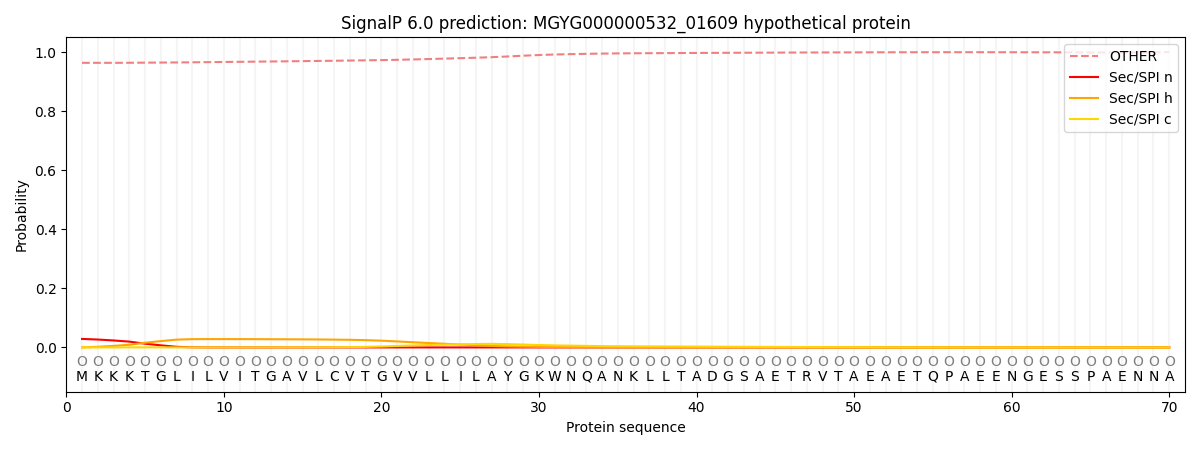
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.965053 | 0.026465 | 0.005194 | 0.000297 | 0.000129 | 0.002882 |