You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000534_00219
You are here: Home > Sequence: MGYG000000534_00219
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
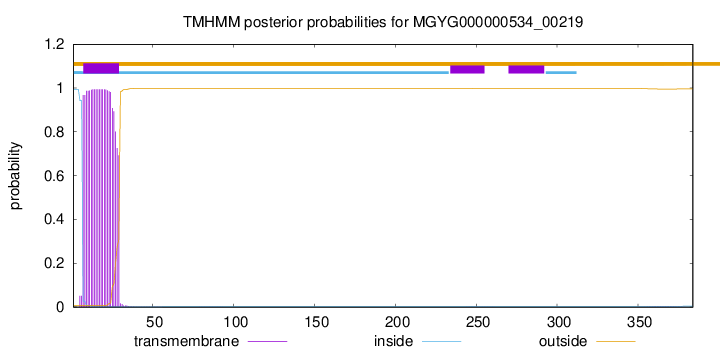
TMHMM annotations
Basic Information help
| Species | Prevotella sp900543975 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900543975 | |||||||||||
| CAZyme ID | MGYG000000534_00219 | |||||||||||
| CAZy Family | GH76 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 91691; End: 92845 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH76 | 48 | 362 | 1.9e-85 | 0.8715083798882681 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam03663 | Glyco_hydro_76 | 1.44e-53 | 40 | 343 | 2 | 315 | Glycosyl hydrolase family 76. Family of alpha-1,6-mannanases. |
| COG4833 | COG4833 | 8.53e-43 | 40 | 381 | 3 | 350 | Predicted alpha-1,6-mannanase, GH76 family [Carbohydrate transport and metabolism]. |
| pfam07470 | Glyco_hydro_88 | 2.52e-10 | 44 | 231 | 2 | 174 | Glycosyl Hydrolase Family 88. Unsaturated glucuronyl hydrolase catalyzes the hydrolytic release of unsaturated glucuronic acids from oligosaccharides (EC:3.2.1.-) produced by the reactions of polysaccharide lyases. |
| COG4225 | YesR | 7.28e-07 | 79 | 233 | 36 | 182 | Rhamnogalacturonyl hydrolase YesR [Carbohydrate transport and metabolism]. |
| cd04791 | LanC_SerThrkinase | 4.45e-06 | 133 | 278 | 148 | 276 | Lanthionine synthetase C-like domain associated with serine/threonine kinases. Some members of this subgroup lack the zinc binding site and the active site residues, and therefore are most likely inactive. The function of this domain is unknown. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QQA30005.1 | 7.31e-152 | 36 | 381 | 31 | 370 |
| BBK86488.1 | 7.31e-152 | 36 | 381 | 31 | 370 |
| QUT65780.1 | 7.31e-152 | 36 | 381 | 31 | 370 |
| QMI79910.1 | 7.31e-152 | 36 | 381 | 31 | 370 |
| QBJ18505.1 | 7.31e-152 | 36 | 381 | 31 | 370 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6SHD_A | 3.42e-74 | 78 | 381 | 88 | 387 | Structureof the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6],6SHD_B Structure of the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6],6SHD_C Structure of the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6] |
| 6Y8F_A | 1.09e-72 | 78 | 381 | 89 | 388 | ChainA, Alpha-1,6-endo-mannanase GH76A mutant [Salegentibacter sp. Hel_I_6] |
| 6SHM_A | 8.55e-72 | 78 | 381 | 89 | 388 | Aninactive (D136A and D137A) variant of alpha-1,6-mannanase, GH76A of Salegentibacter sp. HEL1_6 in complex with alpha-1,6-mannotetrose [Salegentibacter sp. Hel_I_6] |
| 3K7X_A | 1.15e-45 | 40 | 381 | 3 | 338 | ChainA, Lin0763 protein [Listeria innocua] |
| 5M77_A | 1.45e-37 | 30 | 358 | 30 | 352 | ChainA, Alpha-1,6-mannanase [Niallia circulans],5M77_B Chain B, Alpha-1,6-mannanase [Niallia circulans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q75DG6 | 7.20e-11 | 78 | 321 | 65 | 324 | Mannan endo-1,6-alpha-mannosidase DCW1 OS=Ashbya gossypii (strain ATCC 10895 / CBS 109.51 / FGSC 9923 / NRRL Y-1056) OX=284811 GN=DCW1 PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
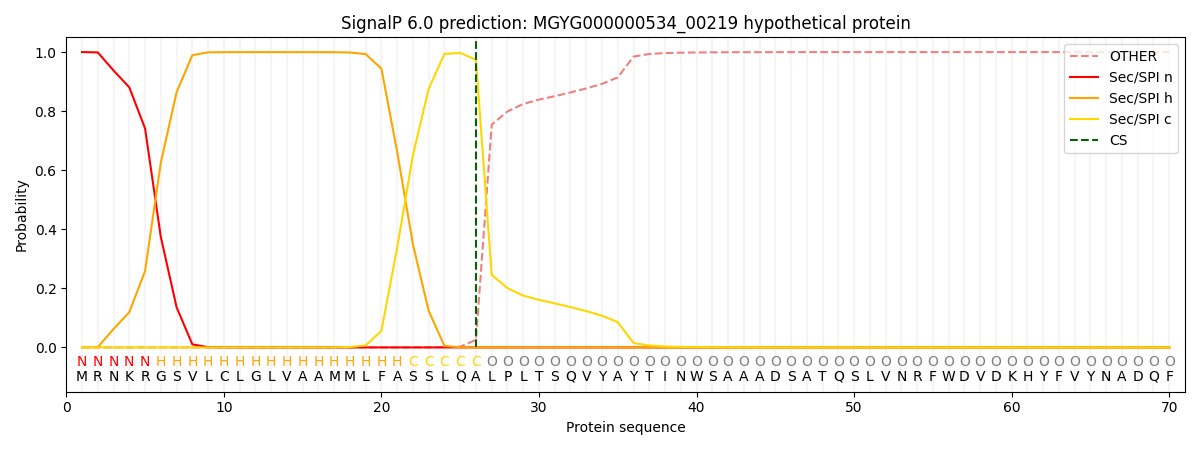
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000368 | 0.998872 | 0.000200 | 0.000195 | 0.000164 | 0.000157 |