You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000537_01874
You are here: Home > Sequence: MGYG000000537_01874
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UMGS1826 sp900555435 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; UMGS1826; UMGS1826 sp900555435 | |||||||||||
| CAZyme ID | MGYG000000537_01874 | |||||||||||
| CAZy Family | GH163 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3453; End: 6065 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH163 | 248 | 478 | 3.1e-31 | 0.8804780876494024 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam16126 | DUF4838 | 1.46e-32 | 251 | 477 | 37 | 259 | Domain of unknown function (DUF4838). This family consists of several uncharacterized proteins found in various Bacteroides and Chloroflexus species. The function of this family is unknown. |
| PRK10150 | PRK10150 | 1.49e-09 | 761 | 848 | 51 | 136 | beta-D-glucuronidase; Provisional |
| pfam02837 | Glyco_hydro_2_N | 5.30e-09 | 760 | 852 | 50 | 138 | Glycosyl hydrolases family 2, sugar binding domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities and has a jelly-roll fold. The domain binds the sugar moiety during the sugar-hydrolysis reaction. |
| PRK05759 | PRK05759 | 7.62e-06 | 512 | 629 | 35 | 135 | F0F1 ATP synthase subunit B; Validated |
| cd06503 | ATP-synt_Fo_b | 4.55e-05 | 522 | 629 | 40 | 130 | F-type ATP synthase, membrane subunit b. Membrane subunit b is a component of the Fo complex of FoF1-ATP synthase. The F-type ATP synthases (FoF1-ATPase) consist of two structural domains: the F1 (assembly factor one) complex containing the soluble catalytic core, and the Fo (oligomycin sensitive factor) complex containing the membrane proton channel, linked together by a central stalk and a peripheral stalk. F1 is composed of alpha (or A), beta (B), gamma (C), delta (D) and epsilon (E) subunits with a stoichiometry of 3:3:1:1:1, while Fo consists of the three subunits a, b, and c (1:2:10-14). An oligomeric ring of 10-14 c subunits (c-ring) make up the Fo rotor. The flux of protons through the ATPase channel (Fo) drives the rotation of the c-ring, which in turn is coupled to the rotation of the F1 complex gamma subunit rotor due to the permanent binding between the gamma and epsilon subunits of F1 and the c-ring of Fo. The F-ATP synthases are primarily found in the inner membranes of eukaryotic mitochondria, in the thylakoid membranes of chloroplasts or in the plasma membranes of bacteria. The F-ATP synthases are the primary producers of ATP, using the proton gradient generated by oxidative phosphorylation (mitochondria) or photosynthesis (chloroplasts). Alternatively, under conditions of low driving force, ATP synthases function as ATPases, thus generating a transmembrane proton or Na(+) gradient at the expense of energy derived from ATP hydrolysis. This group also includes F-ATP synthase that has also been found in the archaea Candidatus Methanoperedens. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNK58589.1 | 6.31e-217 | 33 | 867 | 31 | 769 |
| QNK58588.1 | 9.89e-208 | 31 | 867 | 47 | 784 |
| BBI32737.1 | 2.31e-187 | 36 | 869 | 64 | 785 |
| QNE22644.1 | 2.13e-83 | 118 | 865 | 120 | 758 |
| QDU22041.1 | 3.30e-51 | 21 | 868 | 11 | 743 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6ECA_A | 3.20e-07 | 716 | 848 | 33 | 163 | Lactobacillusrhamnosus Beta-glucuronidase [Lacticaseibacillus rhamnosus],6ECA_B Lactobacillus rhamnosus Beta-glucuronidase [Lacticaseibacillus rhamnosus] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
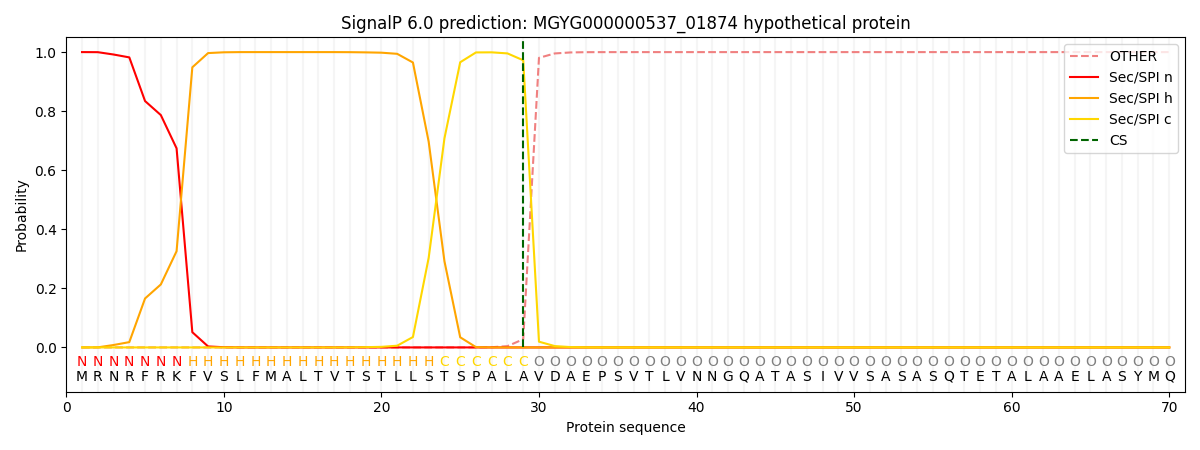
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000263 | 0.999020 | 0.000172 | 0.000215 | 0.000171 | 0.000148 |
