You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000538_01430
You are here: Home > Sequence: MGYG000000538_01430
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UBA1829 sp900549045 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; UBA1829; UBA1829; UBA1829 sp900549045 | |||||||||||
| CAZyme ID | MGYG000000538_01430 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 145423; End: 146898 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 185 | 450 | 6.9e-36 | 0.7418181818181818 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 1.67e-15 | 241 | 446 | 73 | 271 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 2.67e-07 | 235 | 458 | 118 | 374 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM45191.1 | 5.10e-115 | 40 | 487 | 51 | 505 |
| AVM45735.1 | 3.30e-95 | 37 | 487 | 48 | 510 |
| AVM43793.1 | 2.03e-85 | 58 | 485 | 71 | 506 |
| AVM44368.1 | 1.37e-82 | 60 | 485 | 86 | 521 |
| AVM47005.1 | 6.50e-73 | 48 | 485 | 126 | 572 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3NCO_A | 5.11e-12 | 278 | 445 | 119 | 292 | Crystalstructure of FnCel5A from F. nodosum Rt17-B1 [Fervidobacterium nodosum Rt17-B1],3NCO_B Crystal structure of FnCel5A from F. nodosum Rt17-B1 [Fervidobacterium nodosum Rt17-B1] |
| 3RJX_A | 5.11e-12 | 278 | 445 | 119 | 292 | CrystalStructure of Hyperthermophilic Endo-Beta-1,4-glucanase [Fervidobacterium nodosum Rt17-B1] |
| 3RJY_A | 5.11e-12 | 278 | 445 | 119 | 292 | CrystalStructure of Hyperthermophilic Endo-beta-1,4-glucanase in complex with substrate [Fervidobacterium nodosum Rt17-B1] |
| 6UJE_A | 3.01e-10 | 272 | 427 | 103 | 272 | ChainA, Endoglucanase [Clostridioides difficile],6UJF_A Chain A, Endoglucanase [Clostridioides difficile] |
| 3AMC_A | 9.41e-10 | 275 | 445 | 108 | 285 | Crystalstructures of Thermotoga maritima Cel5A, apo form and dimer/au [Thermotoga maritima MSB8],3AMC_B Crystal structures of Thermotoga maritima Cel5A, apo form and dimer/au [Thermotoga maritima MSB8],3AMD_A Crystal structures of Thermotoga maritima Cel5A, apo form and tetramer/au [Thermotoga maritima MSB8],3AMD_B Crystal structures of Thermotoga maritima Cel5A, apo form and tetramer/au [Thermotoga maritima MSB8],3AMD_C Crystal structures of Thermotoga maritima Cel5A, apo form and tetramer/au [Thermotoga maritima MSB8],3AMD_D Crystal structures of Thermotoga maritima Cel5A, apo form and tetramer/au [Thermotoga maritima MSB8],3MMU_A Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_B Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_C Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_D Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_E Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_F Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_G Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_H Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMW_A Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMW_B Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMW_C Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMW_D Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P16169 | 2.44e-09 | 225 | 440 | 64 | 285 | Cellodextrinase A OS=Ruminococcus flavefaciens OX=1265 GN=celA PE=3 SV=3 |
| P14250 | 4.72e-08 | 235 | 370 | 383 | 521 | Endoglucanase 3 OS=Fibrobacter succinogenes (strain ATCC 19169 / S85) OX=59374 GN=cel-3 PE=1 SV=2 |
| P25472 | 5.02e-06 | 232 | 434 | 96 | 284 | Endoglucanase D OS=Ruminiclostridium cellulolyticum (strain ATCC 35319 / DSM 5812 / JCM 6584 / H10) OX=394503 GN=celCCD PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
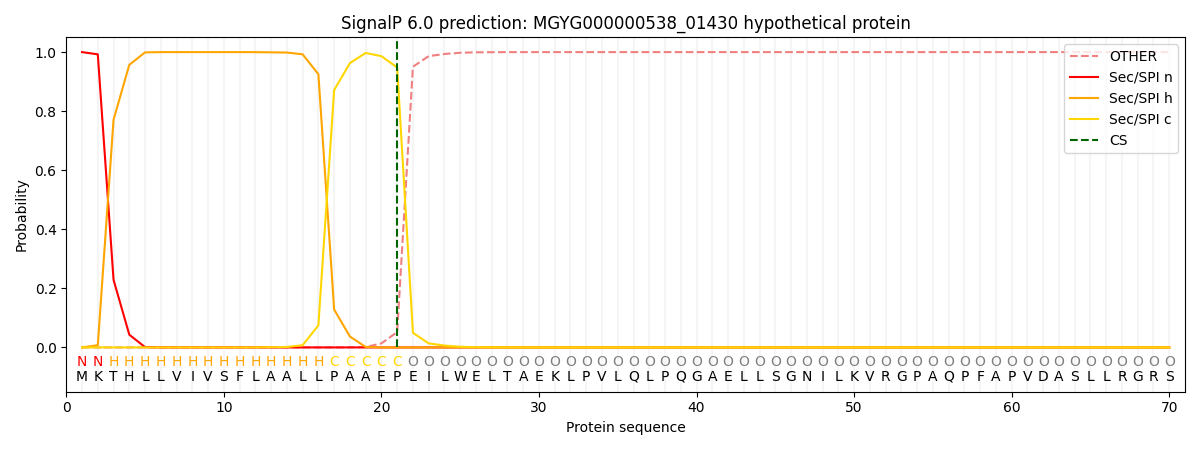
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000231 | 0.999166 | 0.000163 | 0.000148 | 0.000137 | 0.000134 |
