You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000543_01545
You are here: Home > Sequence: MGYG000000543_01545
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Agathobacter sp900552085 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Agathobacter; Agathobacter sp900552085 | |||||||||||
| CAZyme ID | MGYG000000543_01545 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 45967; End: 47265 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 64 | 304 | 8.6e-26 | 0.9580152671755725 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 5.47e-11 | 60 | 278 | 20 | 246 | Cellulase (glycosyl hydrolase family 5). |
| cd00063 | FN3 | 1.93e-04 | 344 | 432 | 2 | 92 | Fibronectin type 3 domain; One of three types of internal repeats found in the plasma protein fibronectin. Its tenth fibronectin type III repeat contains an RGD cell recognition sequence in a flexible loop between 2 strands. Approximately 2% of all animal proteins contain the FN3 repeat; including extracellular and intracellular proteins, membrane spanning cytokine receptors, growth hormone receptors, tyrosine phosphatase receptors, and adhesion molecules. FN3-like domains are also found in bacterial glycosyl hydrolases. |
| smart00060 | FN3 | 8.58e-04 | 349 | 418 | 7 | 81 | Fibronectin type 3 domain. One of three types of internal repeat within the plasma protein, fibronectin. The tenth fibronectin type III repeat contains a RGD cell recognition sequence in a flexible loop between 2 strands. Type III modules are present in both extracellular and intracellular proteins. |
| pfam00041 | fn3 | 0.002 | 349 | 423 | 6 | 85 | Fibronectin type III domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIB28168.1 | 7.53e-143 | 36 | 332 | 4 | 300 |
| QMW91197.1 | 3.27e-137 | 29 | 325 | 21 | 317 |
| BBK76626.1 | 3.27e-137 | 29 | 325 | 21 | 317 |
| QUF84801.1 | 4.63e-137 | 29 | 325 | 21 | 317 |
| AXB85103.1 | 2.64e-136 | 29 | 325 | 21 | 317 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4U5I_A | 1.14e-13 | 20 | 323 | 65 | 374 | ChainA, Endoglucanase H [Acetivibrio thermocellus ATCC 27405],4U5I_B Chain B, Endoglucanase H [Acetivibrio thermocellus ATCC 27405],4U5K_A Chain A, Endoglucanase H [Acetivibrio thermocellus ATCC 27405],4U5K_B Chain B, Endoglucanase H [Acetivibrio thermocellus ATCC 27405] |
| 4U3A_A | 4.82e-13 | 20 | 323 | 65 | 374 | ChainA, Endoglucanase H [Acetivibrio thermocellus ATCC 27405],4U3A_B Chain B, Endoglucanase H [Acetivibrio thermocellus ATCC 27405] |
| 6UJE_A | 3.01e-12 | 65 | 323 | 33 | 305 | ChainA, Endoglucanase [Clostridioides difficile],6UJF_A Chain A, Endoglucanase [Clostridioides difficile] |
| 5BYW_A | 4.98e-11 | 20 | 323 | 65 | 385 | ChainA, Endoglucanase H [Acetivibrio thermocellus ATCC 27405],5BYW_B Chain B, Endoglucanase H [Acetivibrio thermocellus ATCC 27405],5BYW_C Chain C, Endoglucanase H [Acetivibrio thermocellus ATCC 27405],5BYW_D Chain D, Endoglucanase H [Acetivibrio thermocellus ATCC 27405],5BYW_E Chain E, Endoglucanase H [Acetivibrio thermocellus ATCC 27405] |
| 3RJX_A | 1.00e-10 | 78 | 322 | 66 | 313 | CrystalStructure of Hyperthermophilic Endo-Beta-1,4-glucanase [Fervidobacterium nodosum Rt17-B1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P25472 | 1.38e-15 | 1 | 323 | 1 | 324 | Endoglucanase D OS=Ruminiclostridium cellulolyticum (strain ATCC 35319 / DSM 5812 / JCM 6584 / H10) OX=394503 GN=celCCD PE=3 SV=1 |
| P16218 | 5.21e-12 | 20 | 323 | 316 | 625 | Endoglucanase H OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celH PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
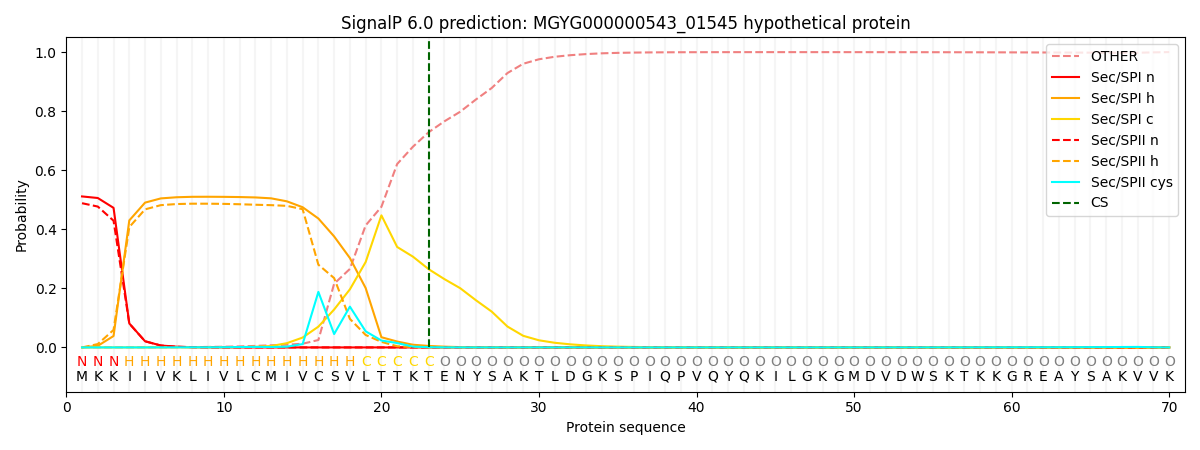
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001259 | 0.501859 | 0.496207 | 0.000206 | 0.000242 | 0.000222 |
