You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000547_01149
You are here: Home > Sequence: MGYG000000547_01149
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
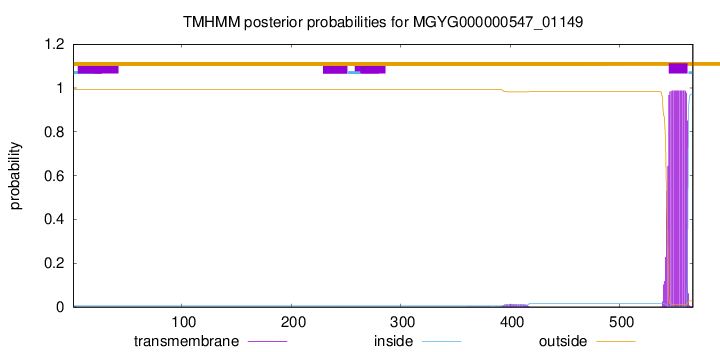
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Erysipelotrichales; Erysipelatoclostridiaceae; Catenibacterium; | |||||||||||
| CAZyme ID | MGYG000000547_01149 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Beta-hexosaminidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 175; End: 1878 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 1 | 146 | 3e-38 | 0.5648148148148148 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1472 | BglX | 1.74e-54 | 1 | 260 | 148 | 367 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| pfam00933 | Glyco_hydro_3 | 2.11e-51 | 1 | 190 | 152 | 316 | Glycosyl hydrolase family 3 N terminal domain. |
| PRK05337 | PRK05337 | 2.13e-27 | 1 | 145 | 147 | 280 | beta-hexosaminidase; Provisional |
| PRK15098 | PRK15098 | 6.46e-17 | 12 | 255 | 201 | 418 | beta-glucosidase BglX. |
| pfam07554 | FIVAR | 0.008 | 444 | 506 | 4 | 69 | FIVAR domain. This domain is found in a wide variety of contexts, but mostly occurring in cell wall associated proteins. A lack of conserved catalytic residues suggests that it is a binding domain. From context, possible substrates are hyaluronate or fibronectin (personal obs: C Yeats). This is further evidenced by. Possibly the exact substrate is N-acetyl glucosamine. Finding it in the same protein as pfam05089 further supports this proposal. It is found in the C-terminal part of Bacillus sp. Gellan lyase, which is removed during maturation. Some of the proteins it is found in are involved in methicillin resistance. The name FIVAR derives from Found In Various Architectures. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCL56675.1 | 2.82e-195 | 1 | 566 | 200 | 795 |
| SQF71406.1 | 1.14e-113 | 1 | 438 | 308 | 738 |
| ABN44479.1 | 1.31e-111 | 1 | 448 | 309 | 749 |
| VDY71833.1 | 6.91e-111 | 1 | 438 | 308 | 738 |
| QKQ43942.1 | 1.03e-110 | 1 | 438 | 311 | 741 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3BMX_A | 2.44e-77 | 8 | 440 | 210 | 638 | Beta-N-hexosaminidase(YbbD) from Bacillus subtilis [Bacillus subtilis],3BMX_B Beta-N-hexosaminidase (YbbD) from Bacillus subtilis [Bacillus subtilis],3NVD_A Structure of YBBD in complex with pugnac [Bacillus subtilis],3NVD_B Structure of YBBD in complex with pugnac [Bacillus subtilis] |
| 3LK6_A | 7.37e-77 | 8 | 440 | 184 | 612 | ChainA, Lipoprotein ybbD [Bacillus subtilis],3LK6_B Chain B, Lipoprotein ybbD [Bacillus subtilis],3LK6_C Chain C, Lipoprotein ybbD [Bacillus subtilis],3LK6_D Chain D, Lipoprotein ybbD [Bacillus subtilis] |
| 4GYJ_A | 1.47e-76 | 8 | 440 | 214 | 642 | Crystalstructure of mutant (D318N) bacillus subtilis family 3 glycoside hydrolase (nagz) in complex with glcnac-murnac (space group P1) [Bacillus subtilis subsp. subtilis str. 168],4GYJ_B Crystal structure of mutant (D318N) bacillus subtilis family 3 glycoside hydrolase (nagz) in complex with glcnac-murnac (space group P1) [Bacillus subtilis subsp. subtilis str. 168],4GYK_A Crystal structure of mutant (D318N) bacillus subtilis family 3 glycoside hydrolase (nagz) in complex with glcnac-murnac (space group P1211) [Bacillus subtilis subsp. subtilis str. 168],4GYK_B Crystal structure of mutant (D318N) bacillus subtilis family 3 glycoside hydrolase (nagz) in complex with glcnac-murnac (space group P1211) [Bacillus subtilis subsp. subtilis str. 168] |
| 4ZM6_A | 8.85e-35 | 1 | 418 | 161 | 531 | Aunique GCN5-related glucosamine N-acetyltransferase region exist in the fungal multi-domain GH3 beta-N-acetylglucosaminidase [Rhizomucor miehei CAU432],4ZM6_B A unique GCN5-related glucosamine N-acetyltransferase region exist in the fungal multi-domain GH3 beta-N-acetylglucosaminidase [Rhizomucor miehei CAU432] |
| 6K5J_A | 8.57e-34 | 1 | 196 | 164 | 342 | Structureof a glycoside hydrolase family 3 beta-N-acetylglucosaminidase from Paenibacillus sp. str. FPU-7 [Paenibacillaceae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P40406 | 1.34e-76 | 8 | 440 | 210 | 638 | Beta-hexosaminidase OS=Bacillus subtilis (strain 168) OX=224308 GN=nagZ PE=1 SV=1 |
| P48823 | 2.27e-43 | 4 | 414 | 193 | 596 | Beta-hexosaminidase A OS=Pseudoalteromonas piscicida OX=43662 GN=cht60 PE=1 SV=1 |
| Q7WUL3 | 4.68e-23 | 1 | 284 | 170 | 435 | Beta-N-acetylglucosaminidase/beta-glucosidase OS=Cellulomonas fimi OX=1708 GN=nag3 PE=1 SV=1 |
| Q7NWB7 | 1.78e-20 | 1 | 144 | 152 | 282 | Beta-hexosaminidase OS=Chromobacterium violaceum (strain ATCC 12472 / DSM 30191 / JCM 1249 / NBRC 12614 / NCIMB 9131 / NCTC 9757) OX=243365 GN=nagZ PE=3 SV=1 |
| Q9JVT3 | 3.39e-20 | 1 | 144 | 156 | 286 | Beta-hexosaminidase OS=Neisseria meningitidis serogroup A / serotype 4A (strain DSM 15465 / Z2491) OX=122587 GN=nagZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
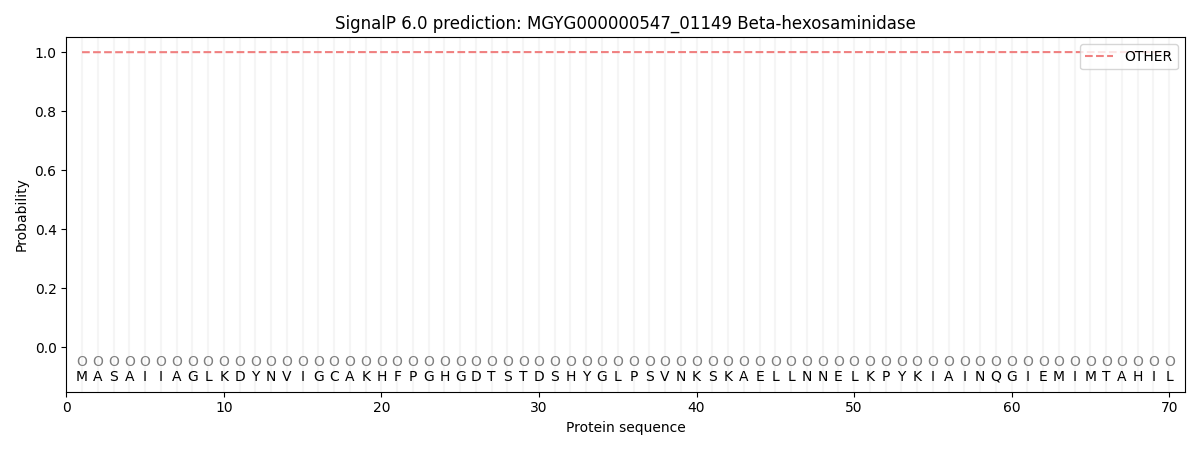
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999714 | 0.000287 | 0.000029 | 0.000001 | 0.000001 | 0.000005 |