You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000553_00592
You are here: Home > Sequence: MGYG000000553_00592
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp900548535 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900548535 | |||||||||||
| CAZyme ID | MGYG000000553_00592 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 118329; End: 120563 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 473 | 734 | 1.9e-43 | 0.6798679867986799 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00633 | Glyco_10 | 4.26e-27 | 474 | 730 | 54 | 260 | Glycosyl hydrolase family 10. |
| pfam00331 | Glyco_hydro_10 | 4.56e-25 | 474 | 735 | 96 | 310 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 5.45e-17 | 481 | 704 | 126 | 304 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam00331 | Glyco_hydro_10 | 9.17e-09 | 69 | 136 | 17 | 85 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 0.001 | 95 | 139 | 1 | 45 | Glycosyl hydrolase family 10. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ASB48786.1 | 7.39e-270 | 21 | 737 | 21 | 732 |
| AVM52702.1 | 2.02e-265 | 17 | 737 | 12 | 746 |
| AVM57878.1 | 8.14e-265 | 17 | 737 | 12 | 746 |
| AXH21505.1 | 1.11e-243 | 2 | 737 | 4 | 743 |
| EDV05072.1 | 1.11e-243 | 2 | 737 | 4 | 743 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4MGS_A | 7.57e-28 | 153 | 297 | 1 | 150 | BiXyn10ACBM1 APO [Bacteroides intestinalis DSM 17393] |
| 4QPW_A | 1.67e-23 | 163 | 294 | 5 | 141 | BiXyn10ACBM1 with Xylohexaose Bound [Bacteroides intestinalis DSM 17393] |
| 4W8L_A | 5.25e-12 | 405 | 717 | 30 | 318 | Structureof GH10 from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4W8L_B Structure of GH10 from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4W8L_C Structure of GH10 from Paenibacillus barcinonensis [Paenibacillus barcinonensis] |
| 4XUY_A | 6.03e-12 | 475 | 737 | 102 | 302 | Crystalstructure of an endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Aspergillus niger [Aspergillus niger CBS 513.88],4XUY_B Crystal structure of an endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Aspergillus niger [Aspergillus niger CBS 513.88] |
| 1US3_A | 1.44e-11 | 495 | 737 | 291 | 513 | Nativexylanase10C from Cellvibrio japonicus [Cellvibrio japonicus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O94163 | 2.20e-13 | 475 | 737 | 125 | 327 | Endo-1,4-beta-xylanase F1 OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=xynF1 PE=1 SV=1 |
| W0HFK8 | 1.32e-12 | 475 | 737 | 131 | 331 | Endo-1,4-beta-xylanase 1 OS=Rhizopus oryzae OX=64495 GN=xyn1 PE=1 SV=1 |
| Q6PRW6 | 9.14e-12 | 475 | 730 | 131 | 324 | Endo-1,4-beta-xylanase OS=Penicillium chrysogenum OX=5076 GN=Xyn PE=1 SV=1 |
| O59859 | 9.80e-12 | 475 | 737 | 127 | 327 | Endo-1,4-beta-xylanase OS=Aspergillus aculeatus OX=5053 GN=xynIA PE=3 SV=1 |
| Q96VB6 | 1.69e-11 | 475 | 737 | 123 | 323 | Endo-1,4-beta-xylanase F3 OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=xynF3 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
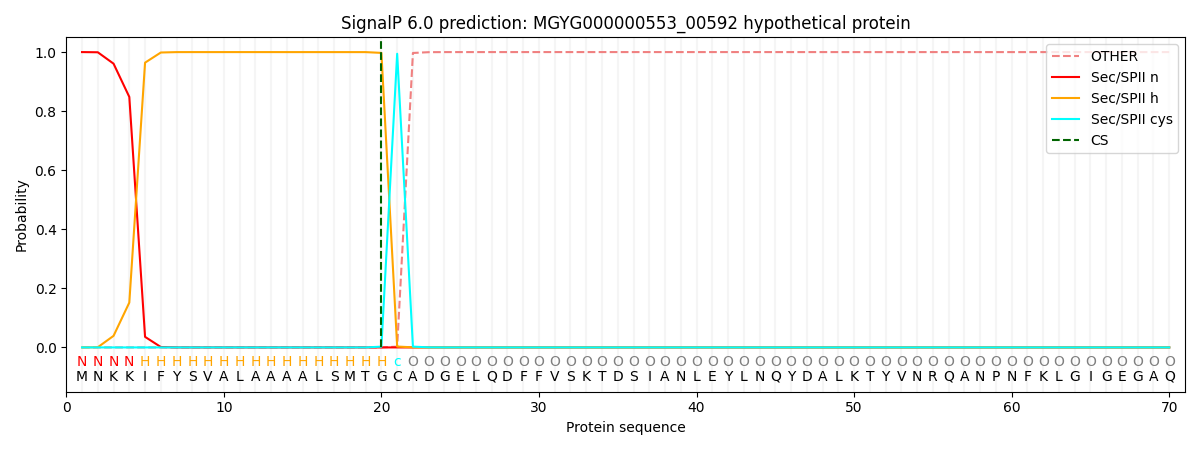
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000050 | 0.000000 | 0.000000 | 0.000000 |
