You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000558_01634
You are here: Home > Sequence: MGYG000000558_01634
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Caecibacter hominis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_C; Negativicutes; Veillonellales; Megasphaeraceae; Caecibacter; Caecibacter hominis | |||||||||||
| CAZyme ID | MGYG000000558_01634 | |||||||||||
| CAZy Family | GT83 | |||||||||||
| CAZyme Description | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 21536; End: 23149 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT83 | 2 | 415 | 2.3e-80 | 0.7851851851851852 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1807 | ArnT | 1.37e-36 | 1 | 434 | 4 | 444 | 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| PRK13279 | arnT | 1.30e-23 | 30 | 338 | 31 | 344 | lipid IV(A) 4-amino-4-deoxy-L-arabinosyltransferase. |
| pfam13231 | PMT_2 | 3.56e-09 | 60 | 219 | 1 | 157 | Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
| COG4745 | COG4745 | 1.24e-04 | 46 | 338 | 46 | 383 | Predicted membrane-bound mannosyltransferase [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AXB83023.1 | 2.17e-271 | 1 | 536 | 1 | 536 |
| AXL21032.1 | 3.98e-218 | 9 | 537 | 9 | 537 |
| ALG41393.1 | 1.31e-191 | 1 | 534 | 1 | 532 |
| AVO26797.1 | 3.74e-191 | 1 | 534 | 1 | 532 |
| CCC72525.1 | 3.74e-191 | 1 | 534 | 1 | 532 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5EZM_A | 4.17e-21 | 27 | 320 | 54 | 356 | CrystalStructure of ArnT from Cupriavidus metallidurans in the apo state [Cupriavidus metallidurans CH34],5F15_A Crystal Structure of ArnT from Cupriavidus metallidurans bound to Undecaprenyl phosphate [Cupriavidus metallidurans CH34] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A4SQX1 | 4.93e-28 | 15 | 337 | 13 | 340 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Aeromonas salmonicida (strain A449) OX=382245 GN=arnT PE=3 SV=1 |
| A0KGY4 | 5.32e-27 | 15 | 337 | 13 | 340 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Aeromonas hydrophila subsp. hydrophila (strain ATCC 7966 / DSM 30187 / BCRC 13018 / CCUG 14551 / JCM 1027 / KCTC 2358 / NCIMB 9240 / NCTC 8049) OX=380703 GN=arnT PE=3 SV=1 |
| O67601 | 1.42e-25 | 9 | 333 | 7 | 335 | Uncharacterized protein aq_1704 OS=Aquifex aeolicus (strain VF5) OX=224324 GN=aq_1704 PE=3 SV=1 |
| B2VBI7 | 4.59e-25 | 16 | 338 | 16 | 343 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Erwinia tasmaniensis (strain DSM 17950 / CFBP 7177 / CIP 109463 / NCPPB 4357 / Et1/99) OX=465817 GN=arnT PE=3 SV=1 |
| C5BDQ8 | 1.97e-24 | 29 | 338 | 29 | 343 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Edwardsiella ictaluri (strain 93-146) OX=634503 GN=arnT PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
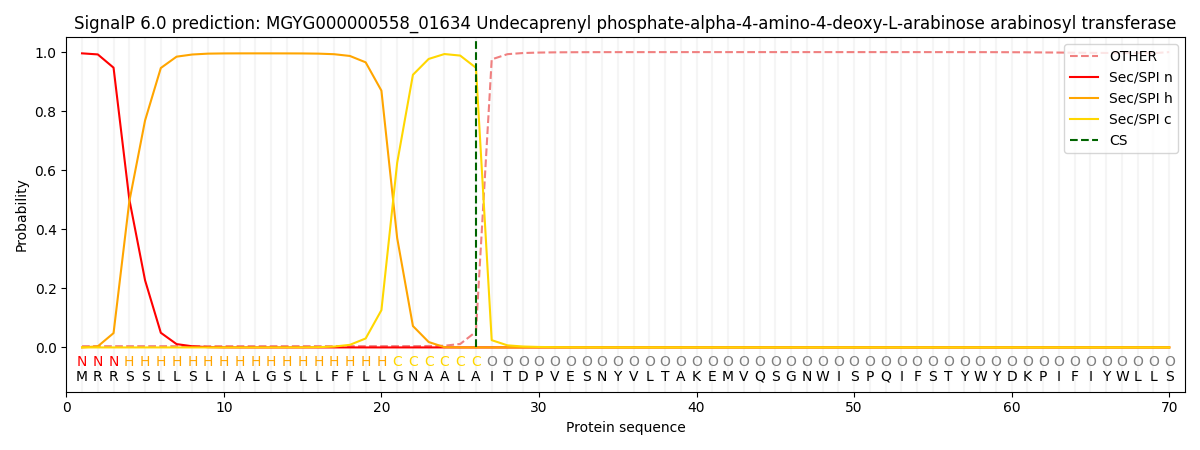
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.017230 | 0.980122 | 0.000637 | 0.000729 | 0.000574 | 0.000694 |
TMHMM Annotations download full data without filtering help
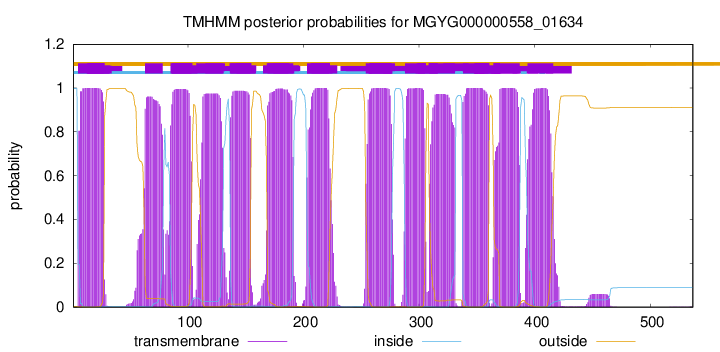
| start | end |
|---|---|
| 5 | 27 |
| 63 | 78 |
| 85 | 103 |
| 108 | 130 |
| 137 | 154 |
| 169 | 191 |
| 203 | 222 |
| 254 | 276 |
| 288 | 305 |
| 309 | 331 |
| 338 | 360 |
| 365 | 387 |
| 394 | 416 |
