You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000559_00964
You are here: Home > Sequence: MGYG000000559_00964
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paraprevotella sp900546665 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Paraprevotella; Paraprevotella sp900546665 | |||||||||||
| CAZyme ID | MGYG000000559_00964 | |||||||||||
| CAZy Family | CBM13 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 50798; End: 53560 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13385 | Laminin_G_3 | 2.52e-26 | 671 | 825 | 1 | 151 | Concanavalin A-like lectin/glucanases superfamily. This domain belongs to the Concanavalin A-like lectin/glucanases superfamily. |
| pfam14200 | RicinB_lectin_2 | 2.98e-13 | 427 | 513 | 3 | 83 | Ricin-type beta-trefoil lectin domain-like. |
| pfam14200 | RicinB_lectin_2 | 2.83e-09 | 478 | 544 | 1 | 73 | Ricin-type beta-trefoil lectin domain-like. |
| TIGR04183 | Por_Secre_tail | 8.94e-08 | 851 | 919 | 5 | 72 | Por secretion system C-terminal sorting domain. Species that include Porphyromonas gingivalis, Fibrobacter succinogenes, Flavobacterium johnsoniae, Cytophaga hutchinsonii, Gramella forsetii, Prevotella intermedia, and Salinibacter ruber average twenty or more copies of a C-terminal domain, represented by this model, associated with sorting to the outer membrane and covalent modification. |
| pfam18962 | Por_Secre_tail | 3.21e-07 | 851 | 918 | 5 | 72 | Secretion system C-terminal sorting domain. Species that include Porphyromonas gingivalis, Fibrobacter succinogenes, Flavobacterium johnsoniae, Cytophaga hutchinsonii, Gramella forsetii, Prevotella intermedia, and Salinibacter ruber have on average twenty or more copies of this C-terminal domain, associated with sorting to the outer membrane and covalent modification. This domain targets proteins to type IX secretion systems and is secreted then cleaved off by a C-terminal signal peptidease. Based on similarity to other families it is likely that this domain adopts an immunoglobulin like fold. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ALJ60208.1 | 0.0 | 2 | 920 | 13 | 940 |
| QUT88791.1 | 0.0 | 3 | 920 | 15 | 940 |
| ASB47762.1 | 5.04e-305 | 20 | 919 | 28 | 932 |
| APZ45922.1 | 1.40e-255 | 22 | 920 | 23 | 930 |
| AUC19784.1 | 1.40e-255 | 22 | 920 | 23 | 930 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4QVS_A | 3.85e-17 | 665 | 832 | 45 | 224 | ChainA, S-layer domain-containing protein [Acetivibrio thermocellus ATCC 27405] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P36905 | 2.08e-07 | 537 | 623 | 927 | 1016 | Amylopullulanase OS=Thermoanaerobacterium saccharolyticum OX=28896 GN=apu PE=3 SV=2 |
| P38536 | 1.95e-06 | 540 | 656 | 932 | 1042 | Amylopullulanase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=amyB PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
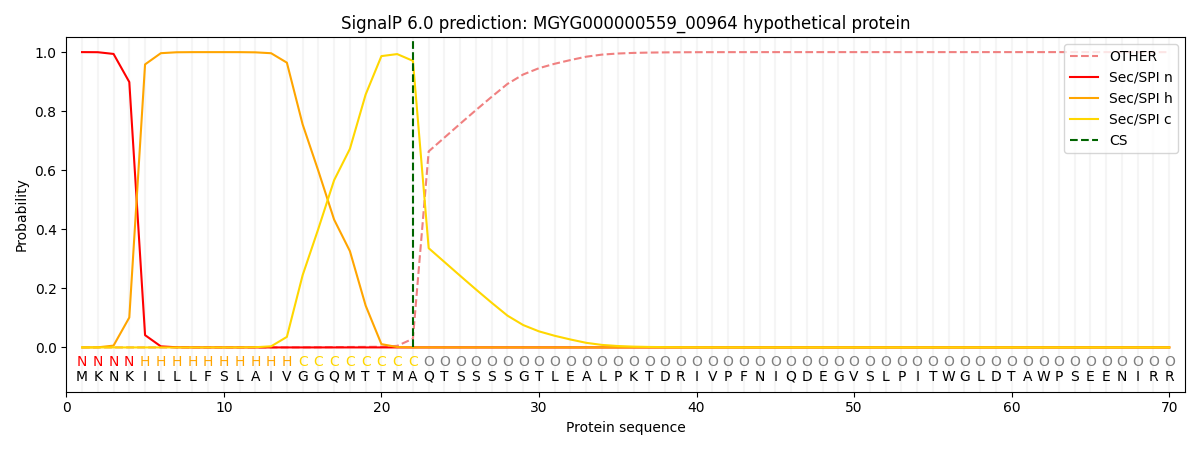
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000572 | 0.998546 | 0.000310 | 0.000208 | 0.000174 | 0.000161 |
