You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000559_02019
You are here: Home > Sequence: MGYG000000559_02019
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paraprevotella sp900546665 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Paraprevotella; Paraprevotella sp900546665 | |||||||||||
| CAZyme ID | MGYG000000559_02019 | |||||||||||
| CAZy Family | GH97 | |||||||||||
| CAZyme Description | Retaining alpha-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 47529; End: 48515 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH97 | 10 | 316 | 2.5e-63 | 0.4849445324881141 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam14508 | GH97_N | 8.73e-56 | 26 | 279 | 1 | 235 | Glycosyl-hydrolase 97 N-terminal. This N-terminal domain of glycosyl-hydrolase-97 contributes part of the active site pocket. It is also important for contact with the catalytic and C-terminal domains of the whole. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADQ79777.1 | 1.36e-108 | 9 | 316 | 9 | 318 |
| QNR85647.1 | 5.85e-105 | 11 | 316 | 12 | 322 |
| QEC79490.1 | 9.90e-103 | 14 | 316 | 19 | 322 |
| AHW58819.1 | 2.42e-96 | 9 | 316 | 11 | 318 |
| AEV98766.1 | 5.43e-95 | 9 | 316 | 7 | 320 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5XFM_A | 1.16e-23 | 20 | 315 | 15 | 319 | Crystalstructure of beta-arabinopyranosidase [Bacteroides thetaiotaomicron],5XFM_B Crystal structure of beta-arabinopyranosidase [Bacteroides thetaiotaomicron],5XFM_C Crystal structure of beta-arabinopyranosidase [Bacteroides thetaiotaomicron],5XFM_D Crystal structure of beta-arabinopyranosidase [Bacteroides thetaiotaomicron] |
| 5HQC_A | 2.51e-10 | 73 | 307 | 42 | 280 | AGlycoside Hydrolase Family 97 enzyme R171K variant from Pseudoalteromonas sp. strain K8 [Pseudoalteromonas sp. K8] |
| 5HQ4_A | 3.34e-10 | 73 | 307 | 42 | 280 | AGlycoside Hydrolase Family 97 enzyme from Pseudoalteromonas sp. strain K8 [Pseudoalteromonas sp. K8],5HQA_A A Glycoside Hydrolase Family 97 enzyme in complex with Acarbose from Pseudoalteromonas sp. strain K8 [Pseudoalteromonas sp. K8] |
| 5HQB_A | 3.34e-10 | 73 | 307 | 42 | 280 | AGlycoside Hydrolase Family 97 enzyme (E480Q) in complex with Panose from Pseudoalteromonas sp. strain K8 [Pseudoalteromonas sp. K8] |
| 2JKA_A | 1.44e-08 | 24 | 309 | 14 | 348 | Nativestructure of a family 97 alpha-glucosidase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482],2JKA_B Native structure of a family 97 alpha-glucosidase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482],2JKE_A Structure of a family 97 alpha-glucosidase from Bacteroides thetaiotaomicron in complex with deoxynojirimycin [Bacteroides thetaiotaomicron VPI-5482],2JKE_B Structure of a family 97 alpha-glucosidase from Bacteroides thetaiotaomicron in complex with deoxynojirimycin [Bacteroides thetaiotaomicron VPI-5482],2JKP_A Structure of a family 97 alpha-glucosidase from Bacteroides thetaiotaomicron in complex with castanospermine [Bacteroides thetaiotaomicron VPI-5482],2JKP_B Structure of a family 97 alpha-glucosidase from Bacteroides thetaiotaomicron in complex with castanospermine [Bacteroides thetaiotaomicron VPI-5482] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| D7CFN7 | 1.04e-16 | 9 | 299 | 20 | 297 | Probable retaining alpha-galactosidase OS=Streptomyces bingchenggensis (strain BCW-1) OX=749414 GN=SBI_01652 PE=3 SV=1 |
| G8JZS4 | 7.94e-08 | 24 | 309 | 25 | 359 | Glucan 1,4-alpha-glucosidase SusB OS=Bacteroides thetaiotaomicron (strain ATCC 29148 / DSM 2079 / JCM 5827 / CCUG 10774 / NCTC 10582 / VPI-5482 / E50) OX=226186 GN=susB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
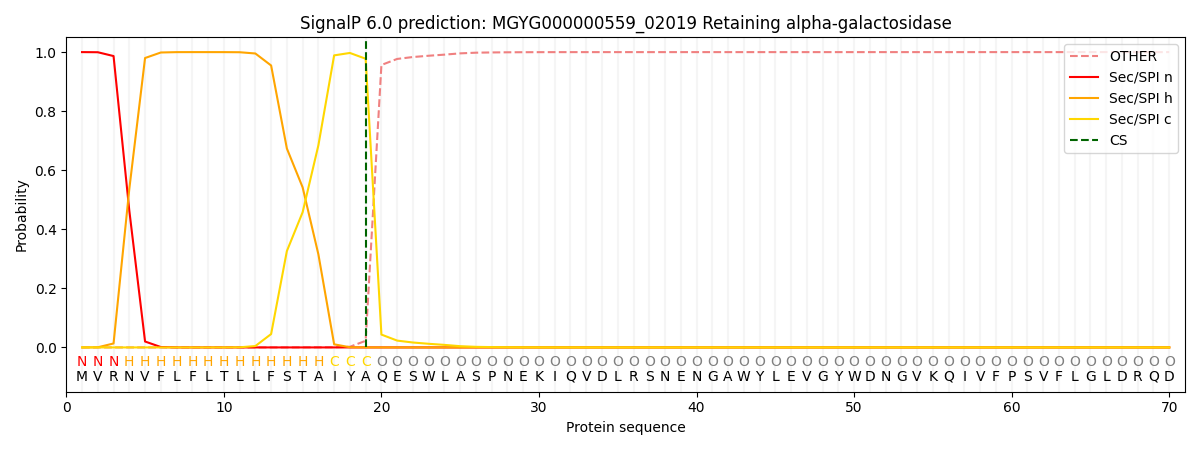
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000469 | 0.998752 | 0.000211 | 0.000187 | 0.000184 | 0.000177 |
