You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000559_02067
You are here: Home > Sequence: MGYG000000559_02067
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paraprevotella sp900546665 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Paraprevotella; Paraprevotella sp900546665 | |||||||||||
| CAZyme ID | MGYG000000559_02067 | |||||||||||
| CAZy Family | GH98 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 977; End: 4129 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH98 | 25 | 342 | 1.1e-83 | 0.9908256880733946 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08306 | Glyco_hydro_98M | 3.00e-86 | 25 | 342 | 5 | 328 | Glycosyl hydrolase family 98. This domain is the putative catalytic domain of glycosyl hydrolase family 98 proteins. |
| pfam08307 | Glyco_hydro_98C | 1.49e-12 | 347 | 585 | 1 | 268 | Glycosyl hydrolase family 98 C-terminal domain. This putative domain is found at the C-terminus of glycosyl hydrolase family 98 proteins. This domain is not expected to form part of the catalytic activity. |
| cd04083 | CBM35_Lmo2446-like | 4.87e-10 | 646 | 748 | 22 | 125 | Carbohydrate Binding Module 35 (CBM35) domains similar to Lmo2446. This family includes carbohydrate binding module 35 (CBM35) domains that are appended to several carbohydrate binding enzymes. Some CBM35 domains belonging to this family are appended to glycoside hydrolase (GH) family domains, including glycoside hydrolase family 31 (GH31), for example the CBM35 domain of Lmo2446, an uncharacterized protein from Listeria monocytogenes EGD-e. These CBM35s are non-catalytic carbohydrate binding domains that facilitate the strong binding of the GH catalytic modules with their dedicated, insoluble substrates. GH31 has a wide range of hydrolytic activities such as alpha-glucosidase, alpha-xylosidase, 6-alpha-glucosyltransferase, or alpha-1,4-glucan lyase, cleaving a terminal carbohydrate moiety from a substrate that may be a starch or a glycoprotein. Most characterized GH31 enzymes are alpha-glucosidases. |
| pfam18998 | Flg_new_2 | 1.35e-05 | 758 | 827 | 1 | 71 | Divergent InlB B-repeat domain. This family of domains are found in bacterial cell surface proteins. They are often found in tandem array. This domain is closely related to pfam09479. |
| cd04086 | CBM35_mannanase-like | 0.002 | 646 | 747 | 21 | 117 | Carbohydrate Binding Module 35 (CBM35); appended to several carbohydrate binding enzymes, including several glycoside hydrolase (GH) family 26 mannanase domains. This family includes carbohydrate binding module 35 (CBM35) domains that are appended to several carbohydrate binding enzymes, including periplasmic component of ABC-type sugar transport system involved in carbohydrate transport and metabolism, and several glycoside hydrolase (GH) domains, including GH26. These CBM6s are non-catalytic carbohydrate binding domains that facilitate the strong binding of the GH catalytic modules with their dedicated, insoluble substrates. Examples of proteins having CMB35s belonging to this family are mannanase A from Clostridium thermocellum (GH26), Man26B from Paenibacillus sp. BME-14 (GH26), and the multifunctional Cel44C-Man26A from Paenibacillus polymyxa GS01 (which has two GH domains, GH44 and GH26). GH26 mainly includes mannan endo-1,4-beta-mannosidase which hydrolyzes 1,4-beta-D-linkages in mannans, galacto-mannans, glucomannans, and galactoglucomannans, but displays little activity towards other plant cell wall polysaccharides. A few proteins belonging to this family have additional CBM3 domains; these CBM3s are not found in the CBM6-CBM35-CBM36_like superfamily. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AGB29515.1 | 0.0 | 6 | 1049 | 1 | 1034 |
| AXY74108.1 | 8.23e-177 | 11 | 750 | 16 | 750 |
| AOZ99817.1 | 2.40e-176 | 24 | 845 | 29 | 831 |
| QUT78600.1 | 1.28e-173 | 28 | 746 | 228 | 934 |
| QUT26887.1 | 1.28e-173 | 28 | 746 | 228 | 934 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2WMH_A | 2.71e-18 | 26 | 584 | 28 | 581 | Crystalstructure of the catalytic module of a family 98 glycoside hydrolase from Streptococcus pneumoniae TIGR4 in complex with the H- disaccharide blood group antigen. [Streptococcus pneumoniae TIGR4] |
| 2WMF_A | 6.25e-18 | 26 | 584 | 28 | 581 | Crystalstructure of the catalytic module of a family 98 glycoside hydrolase from Streptococcus pneumoniae TIGR4 (Sp4GH98) in its native form. [Streptococcus pneumoniae TIGR4] |
| 2WMG_A | 1.90e-17 | 206 | 584 | 198 | 581 | Crystalstructure of the catalytic module of a family 98 glycoside hydrolase from Streptococcus pneumoniae TIGR4 (Sp4GH98) in complex with the LewisY pentasaccharide blood group antigen. [Streptococcus pneumoniae TIGR4] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
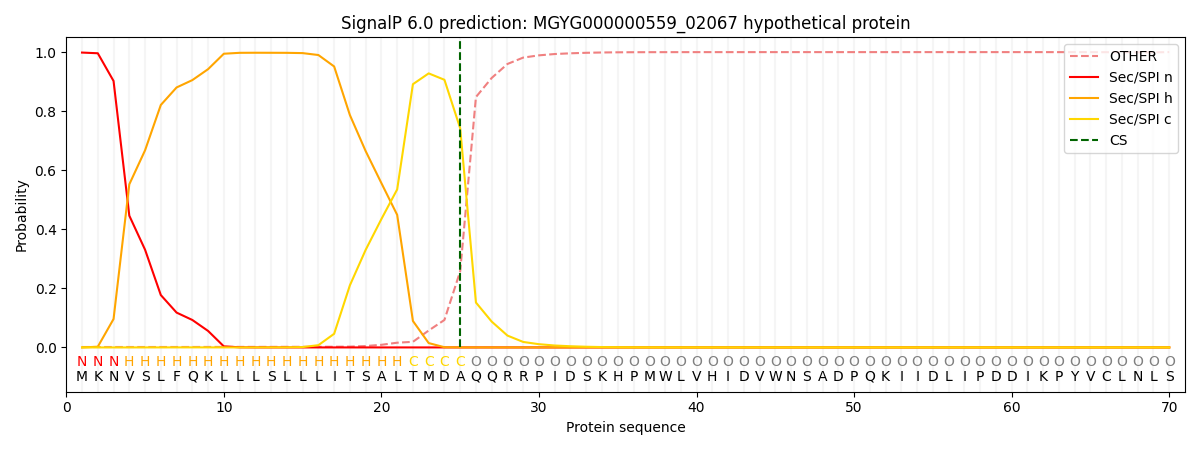
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.003098 | 0.995643 | 0.000434 | 0.000320 | 0.000237 | 0.000214 |
