You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000562_01570
You are here: Home > Sequence: MGYG000000562_01570
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
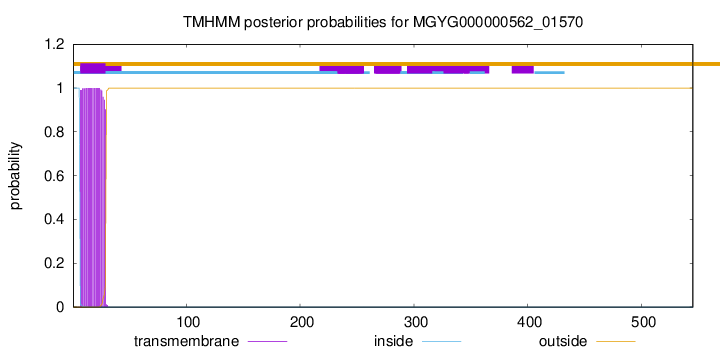
TMHMM annotations
Basic Information help
| Species | Lachnospira sp000437735 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Lachnospira; Lachnospira sp000437735 | |||||||||||
| CAZyme ID | MGYG000000562_01570 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 63909; End: 65546 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 264 | 502 | 5.1e-96 | 0.9831223628691983 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 2.68e-72 | 262 | 511 | 2 | 272 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 3.49e-11 | 271 | 449 | 68 | 227 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| pfam00553 | CBM_2 | 7.92e-05 | 68 | 162 | 1 | 85 | Cellulose binding domain. Two tryptophan residues are involved in cellulose binding. Cellulose binding domain found in bacteria. |
| smart00637 | CBD_II | 1.56e-04 | 87 | 160 | 9 | 76 | CBD_II domain. |
| smart01063 | CBM49 | 0.002 | 76 | 160 | 8 | 82 | Carbohydrate binding domain CBM49. This domain is found at the C terminal of cellulases and in vitro binding studies have shown it to binds to crystalline cellulose. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ACR73731.1 | 9.24e-184 | 1 | 545 | 1 | 535 |
| ADD61840.1 | 2.87e-178 | 1 | 545 | 1 | 538 |
| QTU84139.1 | 2.18e-133 | 250 | 543 | 205 | 498 |
| QWT53734.1 | 1.27e-130 | 59 | 545 | 64 | 514 |
| QNM00780.1 | 1.61e-128 | 56 | 545 | 61 | 514 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6GJF_A | 2.27e-109 | 243 | 544 | 3 | 301 | Ancestralendocellulase Cel5A [synthetic construct],6GJF_B Ancestral endocellulase Cel5A [synthetic construct],6GJF_C Ancestral endocellulase Cel5A [synthetic construct],6GJF_D Ancestral endocellulase Cel5A [synthetic construct],6GJF_E Ancestral endocellulase Cel5A [synthetic construct],6GJF_F Ancestral endocellulase Cel5A [synthetic construct] |
| 4XZB_A | 4.70e-106 | 245 | 543 | 4 | 303 | endo-glucanaseGsCelA P1 [Geobacillus sp. 70PC53] |
| 3PZT_A | 5.77e-105 | 245 | 545 | 29 | 326 | Structureof the endo-1,4-beta-glucanase from Bacillus subtilis 168 with manganese(II) ion [Bacillus subtilis subsp. subtilis str. 168],3PZT_B Structure of the endo-1,4-beta-glucanase from Bacillus subtilis 168 with manganese(II) ion [Bacillus subtilis subsp. subtilis str. 168],3PZU_A P212121 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZU_B P212121 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_A C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_B C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_C C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_D C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168] |
| 4XZW_A | 3.24e-103 | 245 | 543 | 4 | 302 | Endo-glucanasechimera C10 [uncultured bacterium] |
| 1A3H_A | 1.18e-99 | 251 | 544 | 7 | 298 | EndoglucanaseCel5a From Bacillus Agaradherans At 1.6a Resolution [Salipaludibacillus agaradhaerens],2A3H_A Cellobiose Complex Of The Endoglucanase Cel5a From Bacillus Agaradherans At 2.0 A Resolution [Salipaludibacillus agaradhaerens],3A3H_A Cellotriose Complex Of The Endoglucanase Cel5a From Bacillus Agaradherans At 1.6 A Resolution [Salipaludibacillus agaradhaerens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P07983 | 3.43e-102 | 245 | 545 | 34 | 331 | Endoglucanase OS=Bacillus subtilis OX=1423 GN=bglC PE=3 SV=2 |
| P10475 | 6.81e-102 | 245 | 545 | 34 | 331 | Endoglucanase OS=Bacillus subtilis (strain 168) OX=224308 GN=eglS PE=1 SV=1 |
| Q59394 | 5.06e-99 | 245 | 544 | 33 | 329 | Endoglucanase N OS=Pectobacterium atrosepticum OX=29471 GN=celN PE=3 SV=1 |
| P23549 | 6.51e-99 | 245 | 545 | 34 | 331 | Endoglucanase OS=Bacillus subtilis OX=1423 GN=bglC PE=3 SV=1 |
| Q47096 | 1.54e-98 | 245 | 544 | 33 | 329 | Endoglucanase 5 OS=Pectobacterium carotovorum subsp. carotovorum OX=555 GN=celV PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
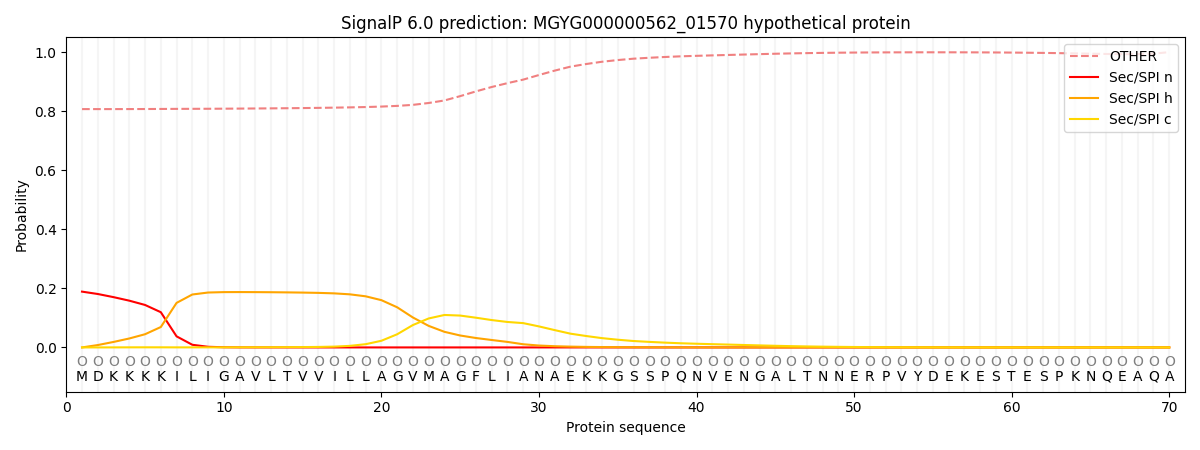
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.816580 | 0.175225 | 0.004025 | 0.001294 | 0.000622 | 0.002263 |