You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000573_00357
You are here: Home > Sequence: MGYG000000573_00357
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
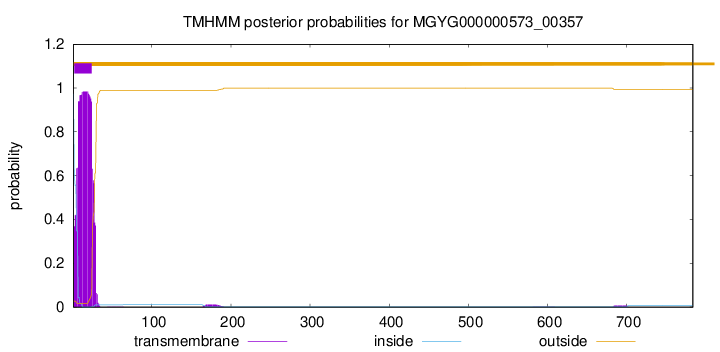
TMHMM annotations
Basic Information help
| Species | Gemmiger variabilis_A | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Gemmiger; Gemmiger variabilis_A | |||||||||||
| CAZyme ID | MGYG000000573_00357 | |||||||||||
| CAZy Family | GT51 | |||||||||||
| CAZyme Description | Penicillin-binding protein 1A | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1098; End: 3452 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT51 | 57 | 235 | 2.3e-57 | 0.9830508474576272 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0744 | MrcB | 6.91e-126 | 4 | 739 | 9 | 606 | Membrane carboxypeptidase (penicillin-binding protein) [Cell wall/membrane/envelope biogenesis]. |
| COG5009 | MrcA | 4.04e-115 | 5 | 767 | 8 | 756 | Membrane carboxypeptidase/penicillin-binding protein [Cell wall/membrane/envelope biogenesis]. |
| pfam00912 | Transgly | 9.56e-68 | 62 | 235 | 6 | 176 | Transglycosylase. The penicillin-binding proteins are bifunctional proteins consisting of transglycosylase and transpeptidase in the N- and C-terminus respectively. The transglycosylase domain catalyzes the polymerization of murein glycan chains. |
| TIGR02071 | PBP_1b | 3.49e-67 | 67 | 766 | 145 | 717 | penicillin-binding protein 1B. Bacterial that synthesize a cell wall of peptidoglycan (murein) generally have several transglycosylases and transpeptidases for the task. This family consists of a particular bifunctional transglycosylase/transpeptidase in E. coli and other Proteobacteria, designated penicillin-binding protein 1B. [Cell envelope, Biosynthesis and degradation of murein sacculus and peptidoglycan] |
| COG4953 | PbpC | 1.03e-52 | 49 | 783 | 36 | 614 | Membrane carboxypeptidase/penicillin-binding protein PbpC [Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBL02306.1 | 2.49e-207 | 2 | 772 | 42 | 794 |
| AXB28952.1 | 1.13e-206 | 2 | 772 | 57 | 809 |
| AXA81016.1 | 1.35e-205 | 2 | 772 | 47 | 799 |
| ATL89188.1 | 4.85e-203 | 9 | 772 | 43 | 784 |
| ATO98888.1 | 7.72e-202 | 9 | 772 | 43 | 784 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7U4H_A | 1.46e-63 | 67 | 782 | 38 | 807 | ChainA, Penicillin-binding protein 1A (Pbp1a) [Chlamydia trachomatis D/UW-3/CX],7U4H_B Chain B, Penicillin-binding protein 1A (Pbp1a) [Chlamydia trachomatis D/UW-3/CX] |
| 4OON_A | 5.63e-49 | 52 | 767 | 23 | 760 | Crystalstructure of PBP1a in complex with compound 17 ((4Z,8S,11E,14S)-5-(2-amino-1,3-thiazol-4-yl)-14-(5,6-dihydroxy-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)-8-formyl-2-methyl-6-oxo-3,10-dioxa-4,7,11-triazatetradeca-4,11-diene-2,12,14-tricarboxylic acid) [Pseudomonas aeruginosa PAO1] |
| 3UDF_A | 5.75e-46 | 39 | 734 | 10 | 720 | ChainA, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDF_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDI_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDI_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDX_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UDX_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE0_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE0_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE1_A Chain A, Penicillin-binding protein 1a [Acinetobacter baumannii],3UE1_B Chain B, Penicillin-binding protein 1a [Acinetobacter baumannii] |
| 3DWK_A | 1.25e-41 | 52 | 731 | 10 | 590 | ChainA, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_B Chain B, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_C Chain C, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL],3DWK_D Chain D, Penicillin-binding protein 2 [Staphylococcus aureus subsp. aureus COL] |
| 2OLU_A | 1.27e-38 | 52 | 769 | 19 | 626 | StructuralInsight Into the Transglycosylation Step Of Bacterial Cell Wall Biosynthesis : Apoenzyme [Staphylococcus aureus],2OLV_A Structural Insight Into the Transglycosylation Step Of Bacterial Cell Wall Biosynthesis : Donor Ligand Complex [Staphylococcus aureus],2OLV_B Structural Insight Into the Transglycosylation Step Of Bacterial Cell Wall Biosynthesis : Donor Ligand Complex [Staphylococcus aureus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8XJ01 | 4.93e-78 | 46 | 746 | 68 | 679 | Penicillin-binding protein 1A OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=pbpA PE=3 SV=1 |
| Q0TNZ8 | 5.57e-75 | 46 | 737 | 68 | 673 | Penicillin-binding protein 1A OS=Clostridium perfringens (strain ATCC 13124 / DSM 756 / JCM 1290 / NCIMB 6125 / NCTC 8237 / Type A) OX=195103 GN=pbpA PE=3 SV=1 |
| Q0SRL7 | 1.63e-73 | 46 | 718 | 68 | 657 | Penicillin-binding protein 1A OS=Clostridium perfringens (strain SM101 / Type A) OX=289380 GN=pbpA PE=3 SV=1 |
| A7GHV1 | 7.37e-70 | 17 | 756 | 29 | 670 | Penicillin-binding protein 1A OS=Clostridium botulinum (strain Langeland / NCTC 10281 / Type F) OX=441772 GN=pbpA PE=3 SV=1 |
| A7FY32 | 9.79e-69 | 17 | 756 | 29 | 670 | Penicillin-binding protein 1A OS=Clostridium botulinum (strain ATCC 19397 / Type A) OX=441770 GN=pbpA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
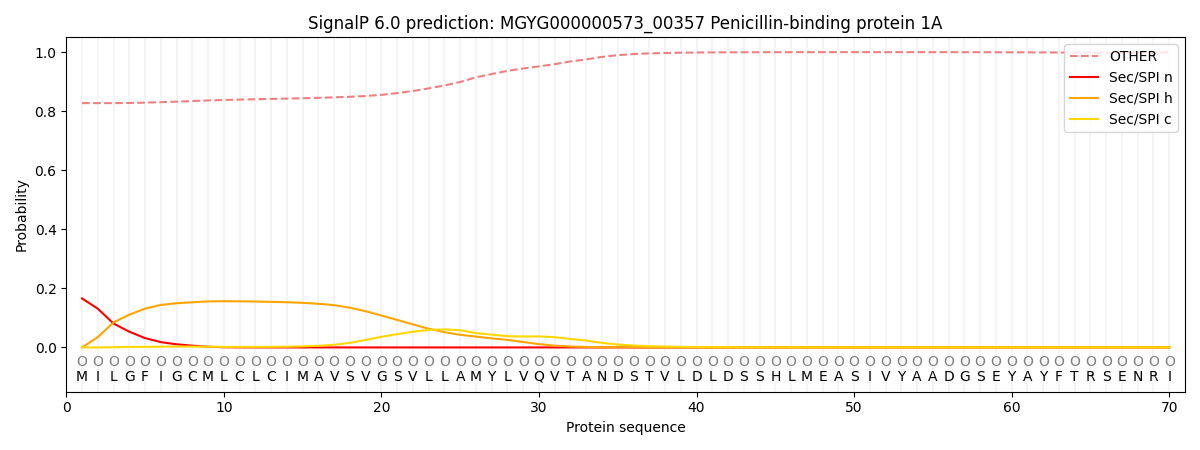
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.836382 | 0.154454 | 0.003574 | 0.000569 | 0.000317 | 0.004709 |