You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000574_00706
You are here: Home > Sequence: MGYG000000574_00706
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
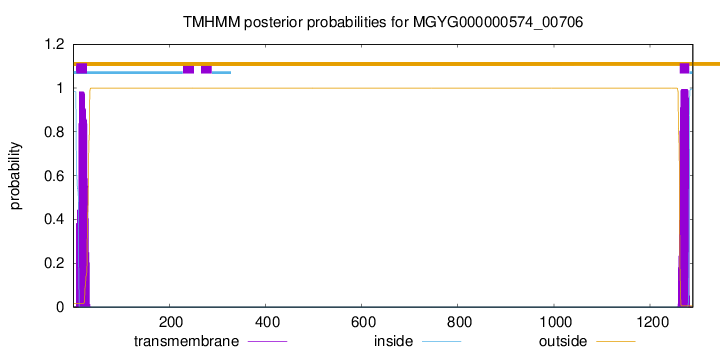
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; ; | |||||||||||
| CAZyme ID | MGYG000000574_00706 | |||||||||||
| CAZy Family | GH136 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2450; End: 6319 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH136 | 457 | 951 | 6.1e-147 | 0.9938900203665988 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam18998 | Flg_new_2 | 2.76e-13 | 171 | 240 | 1 | 74 | Divergent InlB B-repeat domain. This family of domains are found in bacterial cell surface proteins. They are often found in tandem array. This domain is closely related to pfam09479. |
| pfam18998 | Flg_new_2 | 1.17e-06 | 381 | 452 | 7 | 73 | Divergent InlB B-repeat domain. This family of domains are found in bacterial cell surface proteins. They are often found in tandem array. This domain is closely related to pfam09479. |
| pfam13229 | Beta_helix | 1.63e-04 | 638 | 787 | 9 | 144 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| cd00176 | SPEC | 4.11e-04 | 1001 | 1154 | 78 | 213 | Spectrin repeats, found in several proteins involved in cytoskeletal structure; family members include spectrin, alpha-actinin and dystrophin; the spectrin repeat forms a three helix bundle with the second helix interrupted by proline in some sequences; the repeats are independent folding units; tandem repeats are found in differing numbers and arrange in an antiparallel manner to form dimers; the repeats are defined by a characteristic tryptophan (W) residue in helix A and a leucine (L) at the carboxyl end of helix C and separated by a linker of 5 residues; two copies of the repeat are present here |
| pfam13229 | Beta_helix | 4.13e-04 | 539 | 722 | 4 | 140 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUH27719.1 | 6.83e-147 | 459 | 952 | 36 | 508 |
| QXJ59585.1 | 7.33e-108 | 459 | 956 | 35 | 604 |
| AXG37184.1 | 5.52e-94 | 459 | 952 | 35 | 620 |
| BBH21553.1 | 8.75e-88 | 444 | 952 | 28 | 528 |
| QTH41501.1 | 4.79e-86 | 458 | 949 | 329 | 816 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7V6M_A | 6.60e-92 | 455 | 951 | 7 | 576 | ChainA, Fibronectin type III domain-containing protein [Tyzzerella nexilis] |
| 7V6I_A | 5.53e-84 | 459 | 956 | 16 | 613 | ChainA, Lacto-N-biosidase [Bifidobacterium saguini DSM 23967] |
| 5GQC_A | 6.60e-83 | 459 | 951 | 20 | 596 | Crystalstructure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_C Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_D Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_E Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_F Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_G Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_H Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQF_A Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, lacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQF_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, lacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQG_A Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, galacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQG_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, galacto-N-biose complex [Bifidobacterium longum subsp. longum] |
| 6KQT_A | 1.21e-65 | 459 | 826 | 247 | 638 | CrystalStructure of GH136 lacto-N-biosidase from Eubacterium ramulus - native protein [Eubacterium ramulus ATCC 29099] |
| 6KQS_A | 2.72e-63 | 459 | 826 | 247 | 638 | CrystalStructure of GH136 lacto-N-biosidase from Eubacterium ramulus - selenomethionine derivative [Eubacterium ramulus ATCC 29099] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P26831 | 4.51e-11 | 963 | 1157 | 1363 | 1565 | Hyaluronoglucosaminidase OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagH PE=1 SV=2 |
| E8MGH9 | 1.77e-08 | 958 | 1163 | 1666 | 1878 | Beta-L-arabinobiosidase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=hypBA2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
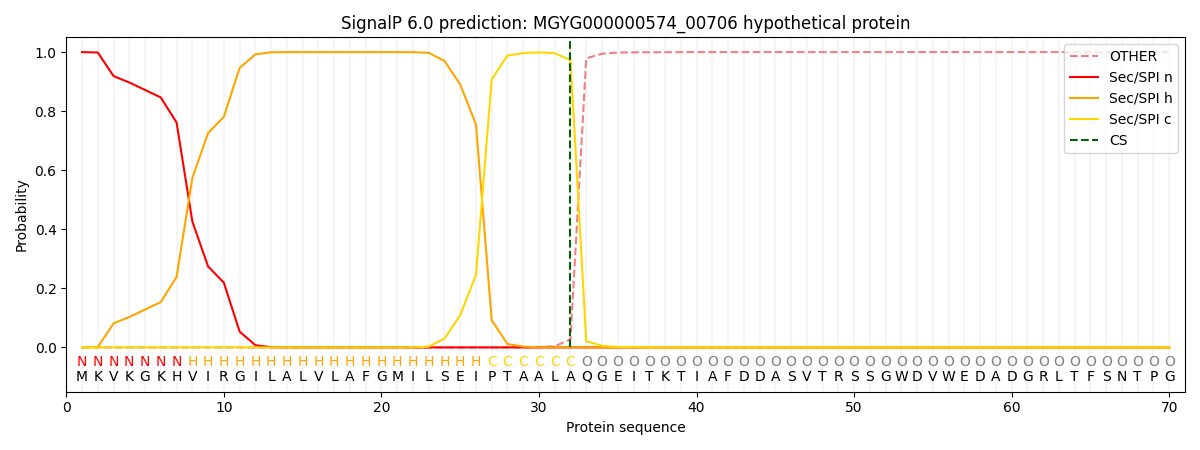
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000226 | 0.999059 | 0.000192 | 0.000199 | 0.000159 | 0.000149 |