You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000574_01264
You are here: Home > Sequence: MGYG000000574_01264
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
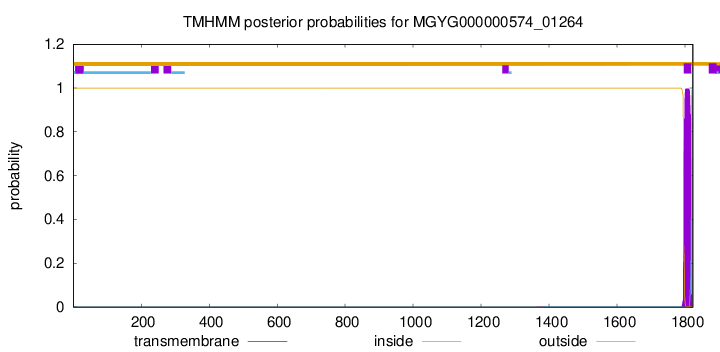
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; ; | |||||||||||
| CAZyme ID | MGYG000000574_01264 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 8; End: 5479 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH95 | 508 | 1172 | 4.2e-48 | 0.8518005540166205 |
| CBM32 | 191 | 338 | 1.9e-18 | 0.9274193548387096 |
| CBM32 | 26 | 160 | 7.7e-18 | 0.8225806451612904 |
| CBM32 | 377 | 506 | 2.2e-17 | 0.8629032258064516 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 4.39e-13 | 381 | 503 | 13 | 124 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam00754 | F5_F8_type_C | 7.99e-11 | 190 | 325 | 1 | 121 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam00754 | F5_F8_type_C | 9.42e-11 | 24 | 157 | 1 | 118 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam17132 | Glyco_hydro_106 | 1.35e-06 | 236 | 342 | 206 | 315 | alpha-L-rhamnosidase. |
| cd00057 | FA58C | 1.60e-06 | 365 | 511 | 14 | 143 | Substituted updates: Jan 31, 2002 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QHW32096.1 | 6.45e-182 | 192 | 1220 | 138 | 1087 |
| QXE33659.1 | 1.01e-143 | 409 | 1214 | 197 | 952 |
| AWS40658.1 | 4.86e-143 | 314 | 1214 | 93 | 926 |
| BCB76148.1 | 3.79e-123 | 314 | 1214 | 49 | 879 |
| QKW17790.1 | 2.02e-79 | 518 | 1190 | 133 | 756 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2XQX_A | 4.36e-10 | 414 | 510 | 56 | 149 | Structureof the family 32 carbohydrate-binding module from Streptococcus pneumoniae EndoD [Streptococcus pneumoniae TIGR4],2XQX_B Structure of the family 32 carbohydrate-binding module from Streptococcus pneumoniae EndoD [Streptococcus pneumoniae TIGR4] |
| 3GDB_A | 4.70e-06 | 414 | 504 | 850 | 937 | Crystalstructure of Spr0440 glycoside hydrolase domain, Endo-D from Streptococcus pneumoniae R6 [Streptococcus pneumoniae R6] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| E8MGH9 | 1.04e-07 | 1623 | 1758 | 1667 | 1806 | Beta-L-arabinobiosidase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=hypBA2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
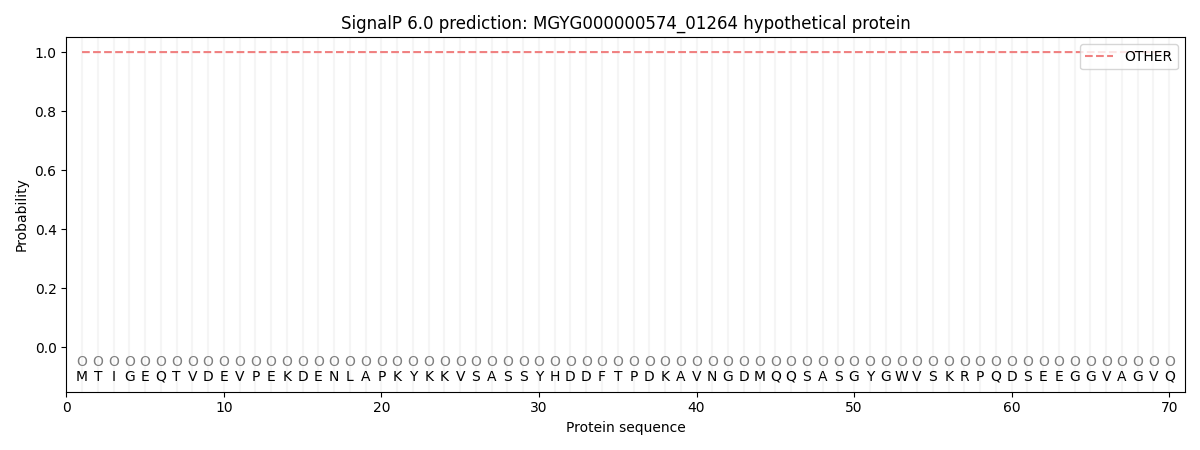
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000038 | 0.000011 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |