You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000575_00591
You are here: Home > Sequence: MGYG000000575_00591
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UBA11452 sp003526375 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; UBA1829; UBA11452; UBA11452 sp003526375 | |||||||||||
| CAZyme ID | MGYG000000575_00591 | |||||||||||
| CAZy Family | GH39 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 11787; End: 13661 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH39 | 73 | 215 | 2.8e-22 | 0.37587006960556846 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3693 | XynA | 1.59e-11 | 63 | 176 | 69 | 191 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| COG2723 | BglB | 8.58e-11 | 42 | 214 | 61 | 263 | Beta-glucosidase/6-phospho-beta-glucosidase/beta-galactosidase [Carbohydrate transport and metabolism]. |
| pfam00331 | Glyco_hydro_10 | 1.79e-09 | 63 | 176 | 46 | 167 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 6.36e-09 | 63 | 176 | 3 | 127 | Glycosyl hydrolase family 10. |
| pfam02449 | Glyco_hydro_42 | 1.20e-06 | 36 | 97 | 6 | 66 | Beta-galactosidase. This group of beta-galactosidase enzymes belong to the glycosyl hydrolase 42 family. The enzyme catalyzes the hydrolysis of terminal, non-reducing terminal beta-D-galactosidase residues. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM44885.1 | 1.51e-133 | 20 | 617 | 21 | 614 |
| QDU57372.1 | 3.79e-80 | 4 | 615 | 7 | 626 |
| AVM44901.1 | 5.18e-51 | 20 | 531 | 19 | 545 |
| AHF90941.1 | 1.74e-36 | 17 | 568 | 220 | 803 |
| QAT16636.1 | 3.93e-34 | 36 | 284 | 113 | 358 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5BX9_A | 4.58e-14 | 29 | 244 | 22 | 252 | Structureof PslG from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],5BXA_A Structure of PslG from Pseudomonas aeruginosa in complex with mannose [Pseudomonas aeruginosa PAO1] |
| 4ZN2_A | 4.58e-14 | 29 | 244 | 22 | 252 | Glycosylhydrolase from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],4ZN2_B Glycosyl hydrolase from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],4ZN2_C Glycosyl hydrolase from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],4ZN2_D Glycosyl hydrolase from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1] |
| 6Z1H_A | 1.50e-10 | 17 | 141 | 47 | 174 | ChainA, ANCESTRAL RECONSTRUCTED GLYCOSIDASE [synthetic construct],6Z1H_B Chain B, ANCESTRAL RECONSTRUCTED GLYCOSIDASE [synthetic construct],6Z1M_A Chain A, Ancestral reconstructed glycosidase [synthetic construct],6Z1M_B Chain B, Ancestral reconstructed glycosidase [synthetic construct],6Z1M_C Chain C, Ancestral reconstructed glycosidase [synthetic construct] |
| 5XGZ_A | 5.32e-08 | 20 | 162 | 44 | 198 | Metagenomicglucose-tolerant glycosidase [uncultured microorganism],5XGZ_B Metagenomic glucose-tolerant glycosidase [uncultured microorganism] |
| 4IPL_A | 9.81e-08 | 39 | 143 | 77 | 192 | Thecrystal structure of 6-phospho-beta-glucosidase BglA-2 from Streptococcus pneumoniae [Streptococcus pneumoniae TIGR4],4IPL_B The crystal structure of 6-phospho-beta-glucosidase BglA-2 from Streptococcus pneumoniae [Streptococcus pneumoniae TIGR4],4IPN_B The complex structure of 6-phospho-beta-glucosidase BglA-2 with thiocellobiose-6P from Streptococcus pneumoniae [Streptococcus pneumoniae TIGR4],4IPN_E The complex structure of 6-phospho-beta-glucosidase BglA-2 with thiocellobiose-6P from Streptococcus pneumoniae [Streptococcus pneumoniae TIGR4] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P42973 | 9.25e-07 | 39 | 140 | 66 | 178 | Aryl-phospho-beta-D-glucosidase BglA OS=Bacillus subtilis (strain 168) OX=224308 GN=bglA PE=1 SV=1 |
| Q7XSK2 | 2.25e-06 | 15 | 141 | 65 | 199 | Beta-glucosidase 16 OS=Oryza sativa subsp. japonica OX=39947 GN=BGLU16 PE=1 SV=2 |
| E3W9M2 | 3.84e-06 | 38 | 187 | 86 | 253 | Cyanidin 3-O-glucoside 5-O-glucosyltransferase (acyl-glucose) OS=Dianthus caryophyllus OX=3570 GN=AA5GT PE=1 SV=1 |
| Q5Z9Z0 | 3.85e-06 | 16 | 138 | 69 | 197 | Beta-glucosidase 24 OS=Oryza sativa subsp. japonica OX=39947 GN=BGLU24 PE=2 SV=1 |
| I1S3C6 | 3.99e-06 | 63 | 139 | 125 | 206 | Endo-1,4-beta-xylanase D OS=Gibberella zeae (strain ATCC MYA-4620 / CBS 123657 / FGSC 9075 / NRRL 31084 / PH-1) OX=229533 GN=XYLD PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
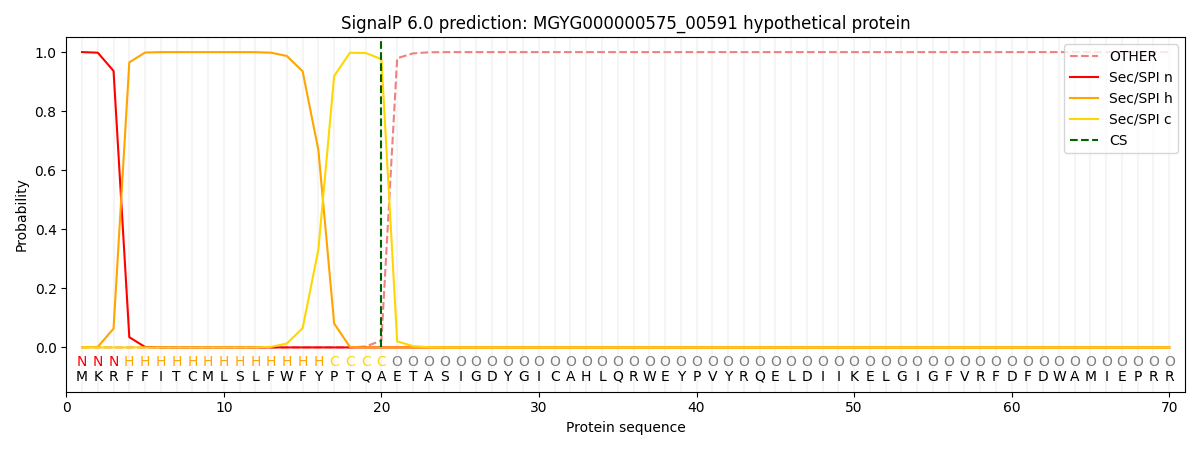
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000375 | 0.998779 | 0.000297 | 0.000180 | 0.000179 | 0.000181 |
