You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000575_01815
You are here: Home > Sequence: MGYG000000575_01815
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UBA11452 sp003526375 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; UBA1829; UBA11452; UBA11452 sp003526375 | |||||||||||
| CAZyme ID | MGYG000000575_01815 | |||||||||||
| CAZy Family | CBM67 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 6846; End: 10148 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM67 | 942 | 1055 | 1.2e-17 | 0.6988636363636364 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02018 | CBM_4_9 | 4.39e-07 | 167 | 291 | 2 | 128 | Carbohydrate binding domain. This family includes diverse carbohydrate binding domains. |
| pfam08531 | Bac_rhamnosid_N | 7.40e-07 | 970 | 1060 | 3 | 114 | Alpha-L-rhamnosidase N-terminal domain. This family consists of bacterial rhamnosidase A and B enzymes. This domain is probably involved in substrate recognition. |
| pfam02018 | CBM_4_9 | 1.05e-04 | 20 | 127 | 2 | 112 | Carbohydrate binding domain. This family includes diverse carbohydrate binding domains. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM43634.1 | 1.14e-118 | 82 | 1079 | 74 | 1067 |
| QNK57364.1 | 4.92e-20 | 414 | 808 | 166 | 541 |
| QZT35774.1 | 1.41e-12 | 540 | 967 | 97 | 521 |
| CCW35746.1 | 9.71e-09 | 943 | 1079 | 24 | 168 |
| QFZ72212.1 | 2.81e-08 | 941 | 1093 | 171 | 338 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6T0Q_A | 7.44e-06 | 942 | 1079 | 198 | 344 | PleurotusOstreatus Lectin (POL), apo form [Pleurotus ostreatus] |
| 6T1D_A | 7.52e-06 | 942 | 1079 | 201 | 347 | PleurotusOstreatus Lectin (POL), compelx with melibiose [Pleurotus ostreatus],6T1D_B Pleurotus Ostreatus Lectin (POL), compelx with melibiose [Pleurotus ostreatus],6T1D_C Pleurotus Ostreatus Lectin (POL), compelx with melibiose [Pleurotus ostreatus],6T1D_D Pleurotus Ostreatus Lectin (POL), compelx with melibiose [Pleurotus ostreatus],6T1D_E Pleurotus Ostreatus Lectin (POL), compelx with melibiose [Pleurotus ostreatus],6T1D_F Pleurotus Ostreatus Lectin (POL), compelx with melibiose [Pleurotus ostreatus] |
| 6KBJ_A | 7.94e-06 | 942 | 1079 | 218 | 364 | Structureof Lectin from Pleurotus ostreatus in complex with malonate [Pleurotus ostreatus],6KBQ_A Crystal Structure of Lectin from Pleurotus ostreatus in complex with Glycerol [Pleurotus ostreatus],6KC2_A Crystal Structure of Lectin from Pleurotus ostreatus in complex with Rhamnose [Pleurotus ostreatus],6LI7_A Crystal Structure of Lectin from Pleurotus ostreatus in complex with GalNAc [Pleurotus ostreatus],6LIK_A Crystal Structure of Lectin from Pleurotus ostreatus in complex with Galactose [Pleurotus ostreatus] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
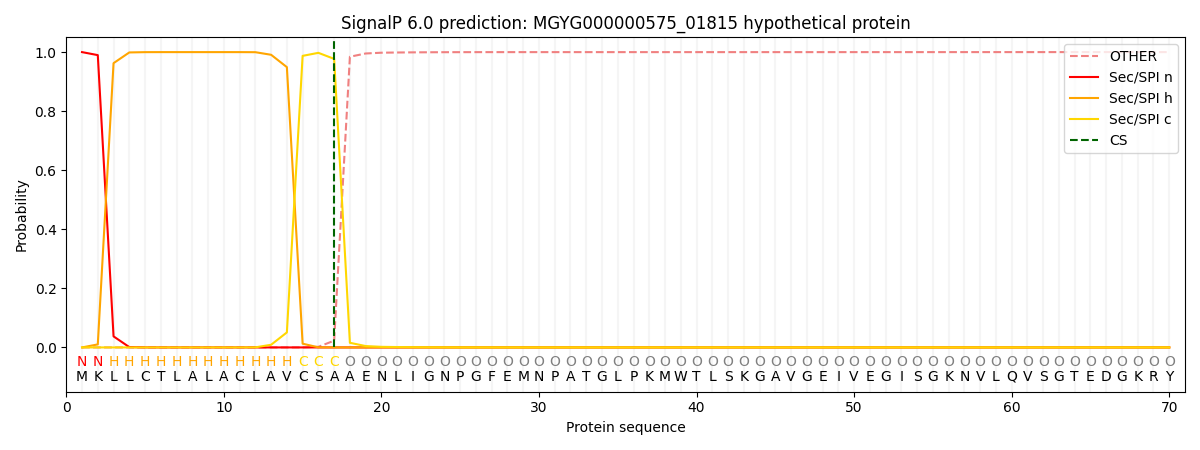
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000200 | 0.999182 | 0.000158 | 0.000144 | 0.000127 | 0.000129 |
