You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000575_01861
Basic Information
help
| Species |
UBA11452 sp003526375
|
| Lineage |
Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; UBA1829; UBA11452; UBA11452 sp003526375
|
| CAZyme ID |
MGYG000000575_01861
|
| CAZy Family |
GH148 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 1070 |
|
119103.58 |
9.2452 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000000575 |
4988607 |
MAG |
Madagascar |
Africa |
|
| Gene Location |
Start: 17597;
End: 20809
Strand: -
|
No EC number prediction in MGYG000000575_01861.
| Family |
Start |
End |
Evalue |
family coverage |
| GH148 |
463 |
610 |
3.7e-30 |
0.993421052631579 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| COG2730
|
BglC |
0.002 |
481 |
636 |
74 |
223 |
Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
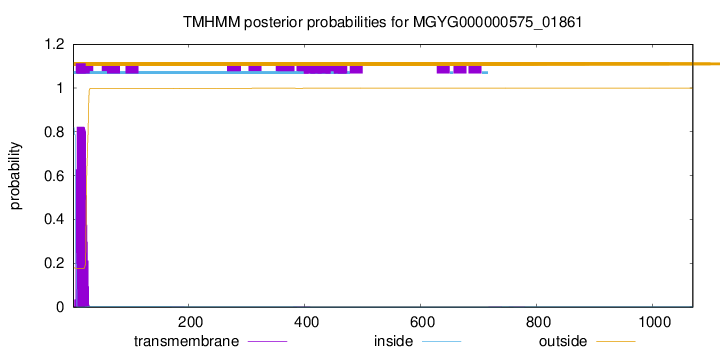
This protein is predicted as SP
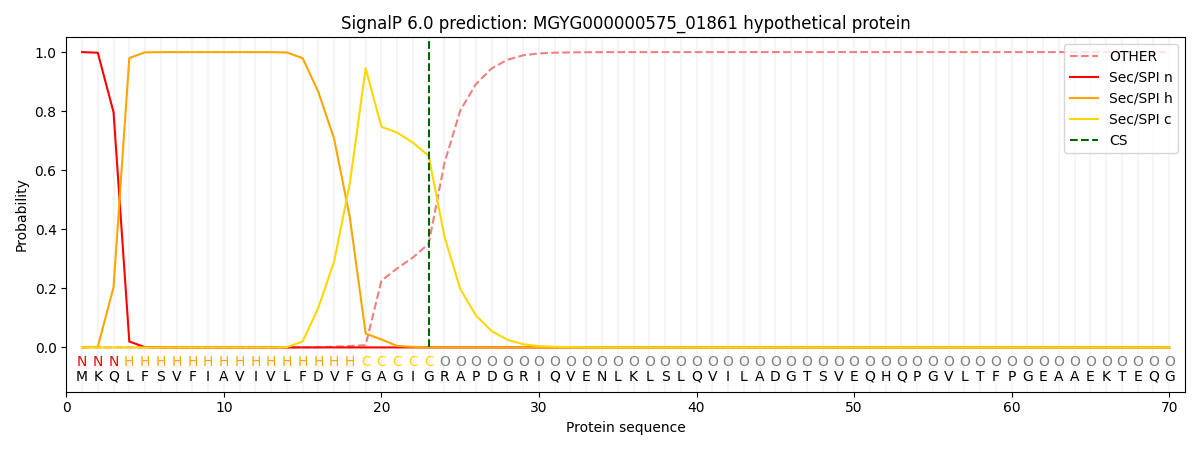
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.000223
|
0.999189
|
0.000160
|
0.000140
|
0.000130
|
0.000126
|