You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000575_01911
You are here: Home > Sequence: MGYG000000575_01911
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
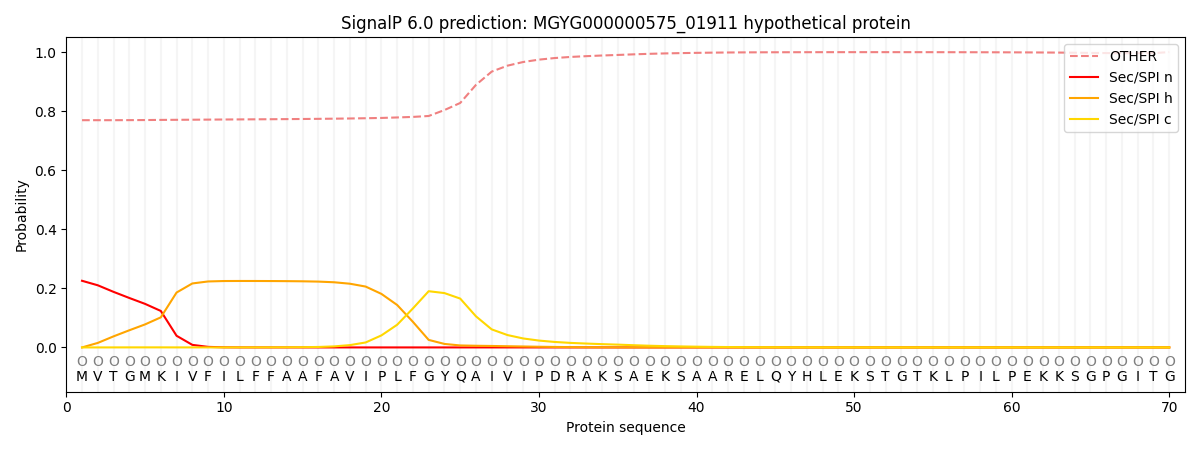
SignalP and Lipop annotations |
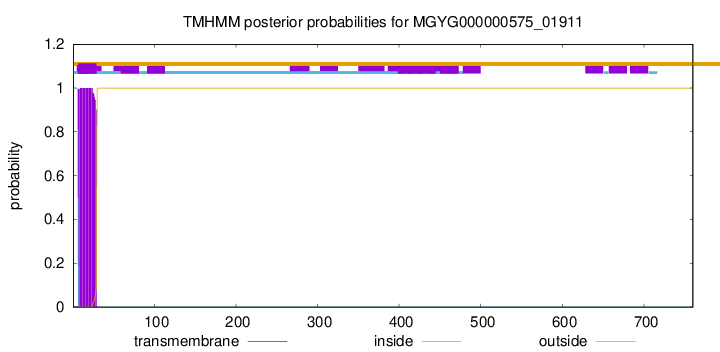
TMHMM annotations
Basic Information help
| Species | UBA11452 sp003526375 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; UBA1829; UBA11452; UBA11452 sp003526375 | |||||||||||
| CAZyme ID | MGYG000000575_01911 | |||||||||||
| CAZy Family | GH163 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 19180; End: 21462 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH163 | 214 | 472 | 5e-29 | 0.9760956175298805 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam16126 | DUF4838 | 9.16e-26 | 206 | 472 | 2 | 262 | Domain of unknown function (DUF4838). This family consists of several uncharacterized proteins found in various Bacteroides and Chloroflexus species. The function of this family is unknown. |
| pfam02837 | Glyco_hydro_2_N | 4.42e-05 | 669 | 746 | 68 | 137 | Glycosyl hydrolases family 2, sugar binding domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities and has a jelly-roll fold. The domain binds the sugar moiety during the sugar-hydrolysis reaction. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QYM78553.1 | 1.03e-104 | 27 | 572 | 55 | 622 |
| QDT41153.1 | 1.35e-51 | 27 | 488 | 593 | 1097 |
| QDT27593.1 | 7.56e-49 | 27 | 479 | 43 | 535 |
| QDV17162.1 | 1.40e-48 | 27 | 479 | 43 | 535 |
| QDU49589.1 | 3.53e-48 | 27 | 479 | 43 | 535 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.780559 | 0.211310 | 0.002385 | 0.000565 | 0.000451 | 0.004732 |