You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000575_02994
You are here: Home > Sequence: MGYG000000575_02994
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UBA11452 sp003526375 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; UBA1829; UBA11452; UBA11452 sp003526375 | |||||||||||
| CAZyme ID | MGYG000000575_02994 | |||||||||||
| CAZy Family | GH39 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 5325; End: 7193 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH39 | 67 | 241 | 2.4e-24 | 0.494199535962877 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00633 | Glyco_10 | 1.12e-07 | 64 | 169 | 3 | 123 | Glycosyl hydrolase family 10. |
| pfam00331 | Glyco_hydro_10 | 6.12e-07 | 64 | 169 | 46 | 163 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 1.16e-06 | 60 | 170 | 65 | 188 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam01229 | Glyco_hydro_39 | 3.13e-04 | 74 | 243 | 74 | 278 | Glycosyl hydrolases family 39. |
| COG1874 | GanA | 0.004 | 29 | 134 | 18 | 121 | Beta-galactosidase GanA [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM44885.1 | 1.49e-126 | 2 | 617 | 4 | 610 |
| QDU57372.1 | 2.06e-77 | 27 | 620 | 28 | 627 |
| AVM44901.1 | 2.85e-62 | 26 | 573 | 24 | 590 |
| ACZ43011.1 | 1.30e-25 | 27 | 244 | 302 | 534 |
| AVM44950.1 | 2.00e-24 | 16 | 240 | 300 | 535 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5JVK_A | 1.09e-10 | 30 | 379 | 107 | 472 | Structuralinsights into a family 39 glycoside hydrolase from the gut symbiont Bacteroides cellulosilyticus WH2. [Bacteroides cellulosilyticus],5JVK_B Structural insights into a family 39 glycoside hydrolase from the gut symbiont Bacteroides cellulosilyticus WH2. [Bacteroides cellulosilyticus],5JVK_C Structural insights into a family 39 glycoside hydrolase from the gut symbiont Bacteroides cellulosilyticus WH2. [Bacteroides cellulosilyticus] |
| 4XUY_A | 6.94e-07 | 52 | 169 | 40 | 168 | Crystalstructure of an endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Aspergillus niger [Aspergillus niger CBS 513.88],4XUY_B Crystal structure of an endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Aspergillus niger [Aspergillus niger CBS 513.88] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A2QFV7 | 4.32e-06 | 52 | 169 | 65 | 193 | Probable endo-1,4-beta-xylanase C OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=xlnC PE=1 SV=1 |
| C5J411 | 4.32e-06 | 52 | 169 | 65 | 193 | Probable endo-1,4-beta-xylanase C OS=Aspergillus niger OX=5061 GN=xlnC PE=2 SV=2 |
| P33559 | 4.32e-06 | 52 | 187 | 65 | 211 | Endo-1,4-beta-xylanase A OS=Aspergillus kawachii (strain NBRC 4308) OX=1033177 GN=xynA PE=1 SV=2 |
| Q96VB6 | 5.64e-06 | 52 | 169 | 61 | 189 | Endo-1,4-beta-xylanase F3 OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=xynF3 PE=1 SV=1 |
| B4XVN1 | 8.32e-06 | 3 | 167 | 16 | 200 | Endo-1,4-beta-xylanase A OS=Streptomyces sp. OX=1931 GN=xynAS9 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
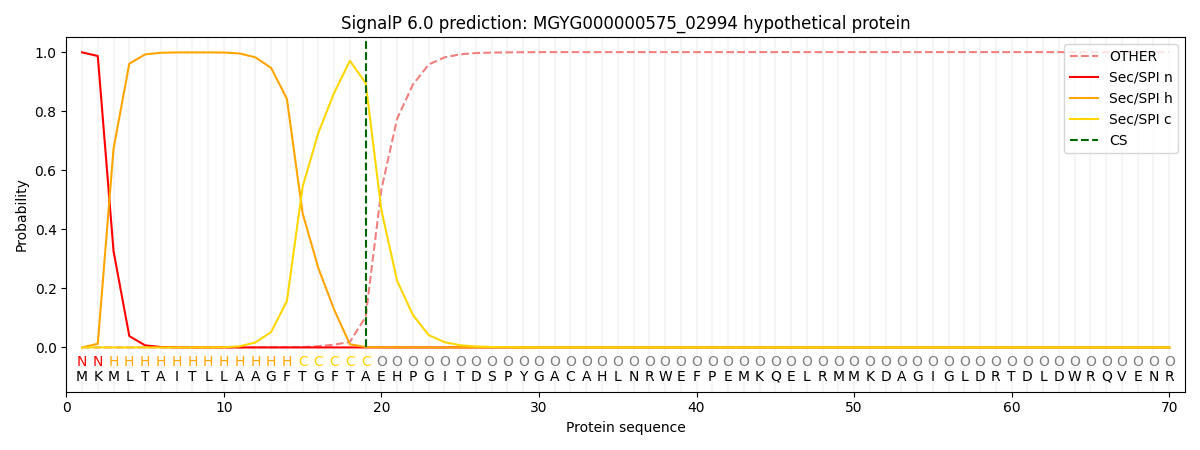
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000974 | 0.996383 | 0.001852 | 0.000257 | 0.000249 | 0.000250 |
