You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000582_00042
You are here: Home > Sequence: MGYG000000582_00042
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
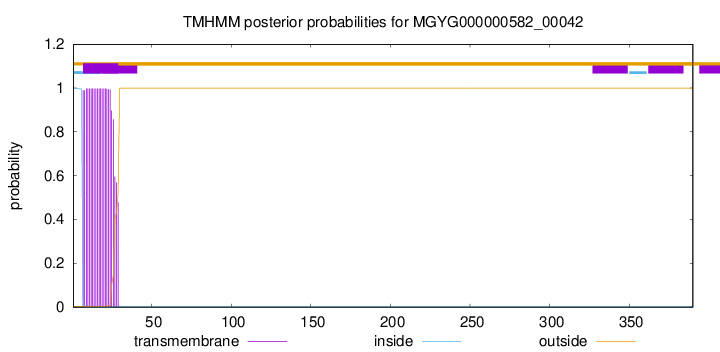
TMHMM annotations
Basic Information help
| Species | Lactococcus garvieae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Streptococcaceae; Lactococcus; Lactococcus garvieae | |||||||||||
| CAZyme ID | MGYG000000582_00042 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | Endo-beta-N-acetylglucosaminidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 42206; End: 43378 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd06542 | GH18_EndoS-like | 3.25e-49 | 36 | 297 | 2 | 247 | Endo-beta-N-acetylglucosaminidases are bacterial chitinases that hydrolyze the chitin core of various asparagine (N)-linked glycans and glycoproteins. The endo-beta-N-acetylglucosaminidases have a glycosyl hydrolase family 18 (GH18) catalytic domain. Some members also have an additional C-terminal glycosyl hydrolase family 20 (GH20) domain while others have an N-terminal domain of unknown function (pfam08522). Members of this family include endo-beta-N-acetylglucosaminidase S (EndoS) from Streptococcus pyogenes, EndoF1, EndoF2, EndoF3, and EndoH from Flavobacterium meningosepticum, and EndoE from Enterococcus faecalis. EndoS is a secreted endoglycosidase from Streptococcus pyogenes that specifically hydrolyzes the glycan on human IgG between two core N-acetylglucosamine residues. EndoE is a secreted endoglycosidase, encoded by the ndoE gene in Enterococcus faecalis, that hydrolyzes the glycan on human RNase B. |
| cd02878 | GH18_zymocin_alpha | 0.007 | 135 | 167 | 93 | 123 | Zymocin, alpha subunit. Zymocin is a heterotrimeric enzyme that inhibits yeast cell cycle progression. The zymocin alpha subunit has a chitinase activity that is essential for holoenzyme action from the cell exterior while the gamma subunit contains the intracellular toxin responsible for G1 phase cell cycle arrest. The zymocin alpha and beta subunits are thought to act from the cell's exterior by docking to the cell wall-associated chitin, thus mediating gamma-toxin translocation. The alpha subunit has an eight-stranded TIM barrel fold similar to that of family 18 glycosyl hydrolases such as hevamine, chitolectin, and chitobiase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BAK60413.1 | 7.27e-287 | 1 | 390 | 1 | 390 |
| BAK58445.1 | 7.27e-287 | 1 | 390 | 1 | 390 |
| QSQ99389.1 | 7.27e-287 | 1 | 390 | 1 | 390 |
| QSR13406.1 | 8.35e-277 | 1 | 390 | 1 | 390 |
| QSR01276.1 | 1.97e-275 | 1 | 390 | 1 | 390 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7PUJ_A | 1.14e-150 | 30 | 389 | 6 | 365 | ChainA, Beta-N-acetylhexosaminidase [Enterococcus faecalis],7PUK_A Chain A, Beta-N-acetylhexosaminidase [Enterococcus faecalis],7PUK_C Chain C, Beta-N-acetylhexosaminidase [Enterococcus faecalis] |
| 6KPL_A | 1.20e-37 | 40 | 315 | 35 | 292 | CrystalStructure of endo-beta-N-acetylglucosaminidase from Cordyceps militaris in apo form [Cordyceps militaris CM01],6KPM_A Crystal Structure of endo-beta-N-acetylglucosaminidase from Cordyceps militaris in complex with L-fucose [Cordyceps militaris CM01] |
| 6KPN_A | 1.72e-36 | 40 | 315 | 35 | 292 | CrystalStructure of endo-beta-N-acetylglucosaminidase from Cordyceps militaris D154N/E156Q mutant in complex with fucosyl-N-acetylglucosamine [Cordyceps militaris CM01],6KPO_A Crystal Structure of endo-beta-N-acetylglucosaminidase from Cordyceps militaris D154N/E156Q mutant in complex with fucosyl-N-acetylglucosamine-Asn [Cordyceps militaris CM01] |
| 4NUY_A | 1.24e-13 | 40 | 292 | 20 | 311 | Crystalstructure of EndoS, an endo-beta-N-acetyl-glucosaminidase from Streptococcus pyogenes [Streptococcus pyogenes serotype M1] |
| 4NUZ_A | 6.87e-13 | 40 | 292 | 20 | 311 | Crystalstructure of a glycosynthase mutant (D233Q) of EndoS, an endo-beta-N-acetyl-glucosaminidase from Streptococcus pyogenes [Streptococcus pyogenes serotype M1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B7GPC7 | 2.36e-121 | 38 | 389 | 54 | 418 | Endo-beta-N-acetylglucosaminidase OS=Bifidobacterium longum subsp. infantis (strain ATCC 15697 / DSM 20088 / JCM 1222 / NCTC 11817 / S12) OX=391904 GN=Blon_2468 PE=1 SV=1 |
| P36912 | 7.04e-32 | 28 | 315 | 54 | 330 | Endo-beta-N-acetylglucosaminidase F2 OS=Elizabethkingia meningoseptica OX=238 GN=endOF2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
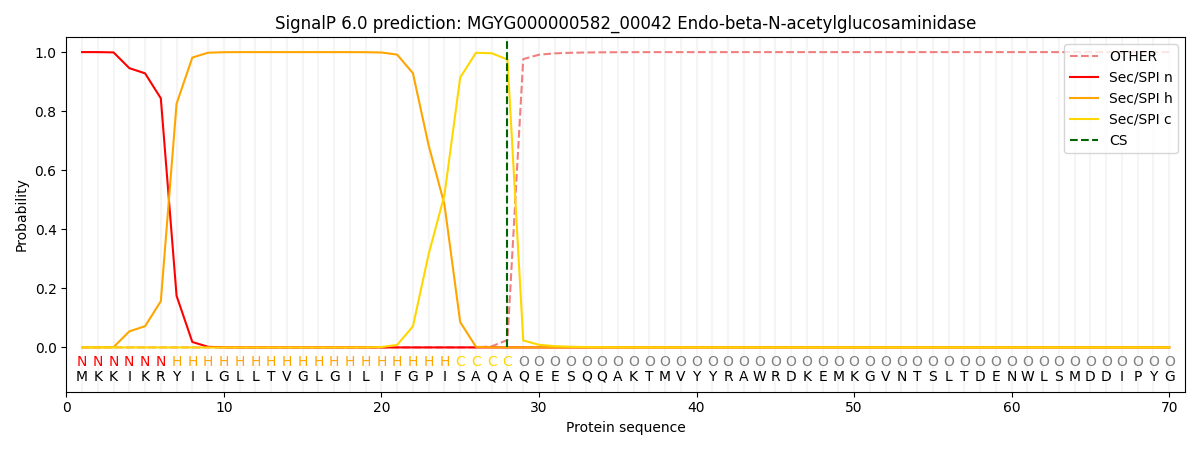
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000334 | 0.998799 | 0.000260 | 0.000219 | 0.000188 | 0.000172 |