You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000582_01156
You are here: Home > Sequence: MGYG000000582_01156
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Lactococcus garvieae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Streptococcaceae; Lactococcus; Lactococcus garvieae | |||||||||||
| CAZyme ID | MGYG000000582_01156 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 26990; End: 27571 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08230 | CW_7 | 8.87e-16 | 34 | 73 | 1 | 40 | CW_7 repeat. This domain was originally found in the C-terminal moiety of the Cpl-7 lysozyme encoded by the Streptococcus pneumoniae bacteriophage Cp-7. It is also found in the cell wall hydrolases of human and life-stock pathogens. CW_7 repeats make up a cell wall binding motif. |
| cd13925 | RPF | 2.53e-13 | 134 | 192 | 1 | 71 | core lysozyme-like domain of resuscitation-promoting factor proteins. Resuscitation-promoting factor (RPF) proteins, found in various (G+C)-rich Gram-positive bacteria, act to reactivate cultures from stationary phase. This protein shares elements of the structural core of lysozyme and related proteins. Furthermore, it shares a conserved active site glutamate which is required for activity, and has a polysaccharide binding cleft that corresponds to the peptidoglycan binding cleft of lysozyme. Muralytic activity of Rpf in Micrococcus luteus correlates with resuscitation, supporting a mechanism dependent on cleavage of peptidoglycan by RPF. |
| smart01095 | Cpl-7 | 5.72e-12 | 33 | 73 | 1 | 41 | Cpl-7 lysozyme C-terminal domain. This domain was originally found in the C-terminal moiety of the Cpl-7 lysozyme encoded by the Streptococcus pneumoniae bacteriophage Cp-7. It is assumed that these repeats represent cell wall binding motifs although no direct evidence has been obtained so far. |
| pfam00062 | Lys | 0.003 | 130 | 163 | 22 | 57 | C-type lysozyme/alpha-lactalbumin family. Alpha-lactalbumin is the regulatory subunit of lactose synthase, changing the substrate specificity of galactosyltransferase from N-acetylglucosamine to glucose. C-type lysozymes are secreted bacteriolytic enzymes that cleave the peptidoglycan of bacterial cell walls. Structure is a multi-domain, mixed alpha and beta fold, containing four conserved disulfide bonds. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QSQ99095.1 | 2.86e-133 | 1 | 193 | 1 | 193 |
| QSR01029.1 | 2.86e-133 | 1 | 193 | 1 | 193 |
| QSR03502.1 | 1.17e-132 | 1 | 193 | 1 | 193 |
| QSR11664.1 | 1.17e-132 | 1 | 193 | 1 | 193 |
| QSR13341.1 | 1.17e-132 | 1 | 193 | 1 | 193 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q4A0G5 | 1.92e-16 | 113 | 192 | 164 | 243 | Probable transglycosylase IsaA OS=Staphylococcus saprophyticus subsp. saprophyticus (strain ATCC 15305 / DSM 20229 / NCIMB 8711 / NCTC 7292 / S-41) OX=342451 GN=isaA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
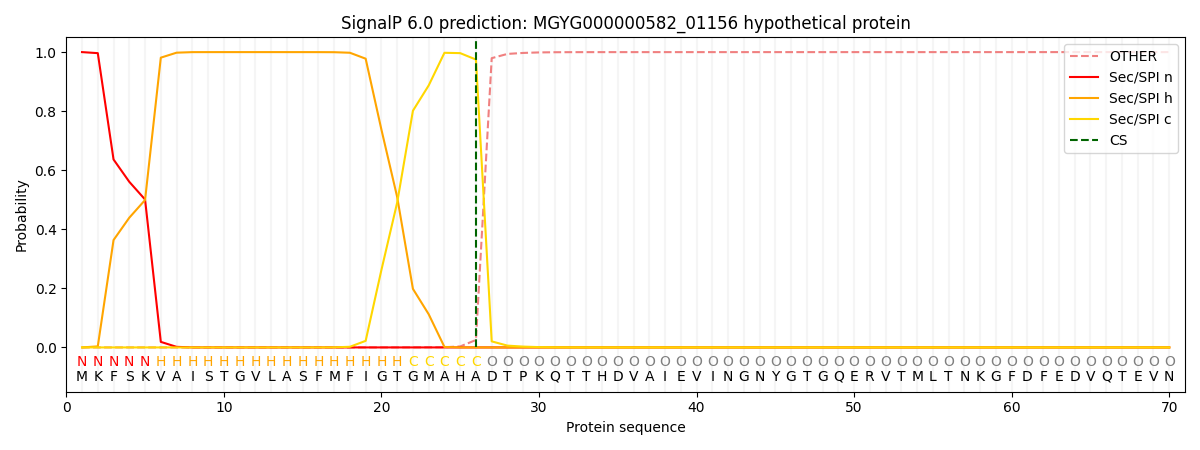
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000252 | 0.999018 | 0.000198 | 0.000184 | 0.000174 | 0.000153 |
