You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000583_01017
You are here: Home > Sequence: MGYG000000583_01017
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
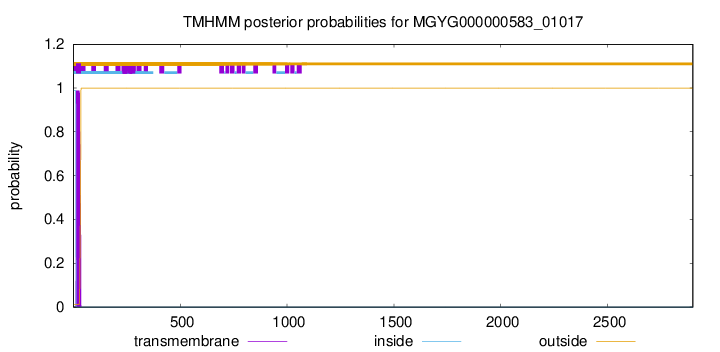
TMHMM annotations
Basic Information help
| Species | Acetatifactor sp900766575 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Acetatifactor; Acetatifactor sp900766575 | |||||||||||
| CAZyme ID | MGYG000000583_01017 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 34262; End: 42961 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL9 | 2201 | 2592 | 2e-121 | 0.976 |
| CE8 | 993 | 1332 | 2.8e-43 | 0.9444444444444444 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PLN02682 | PLN02682 | 1.00e-15 | 994 | 1274 | 76 | 289 | pectinesterase family protein |
| PLN02665 | PLN02665 | 2.13e-14 | 992 | 1274 | 69 | 282 | pectinesterase family protein |
| PLN02432 | PLN02432 | 3.07e-14 | 996 | 1276 | 19 | 216 | putative pectinesterase |
| PLN02217 | PLN02217 | 9.37e-14 | 1136 | 1274 | 328 | 470 | probable pectinesterase/pectinesterase inhibitor |
| COG4677 | PemB | 4.36e-13 | 997 | 1275 | 90 | 332 | Pectin methylesterase and related acyl-CoA thioesterases [Carbohydrate transport and metabolism, Lipid transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ABX44216.1 | 0.0 | 1475 | 2607 | 82 | 1197 |
| BCJ94249.1 | 7.54e-305 | 1563 | 2839 | 159 | 1445 |
| ADL35206.1 | 6.16e-279 | 969 | 2613 | 693 | 2465 |
| ACR71158.1 | 2.14e-227 | 1627 | 2600 | 617 | 1644 |
| AKX86292.1 | 2.91e-177 | 1811 | 2616 | 68 | 859 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1RU4_A | 1.45e-39 | 2202 | 2494 | 15 | 301 | ChainA, Pectate lyase [Dickeya chrysanthemi] |
| 5OLQ_A | 3.47e-19 | 2202 | 2488 | 3 | 322 | Rhamnogalacturonanlyase [Bacteroides thetaiotaomicron],5OLQ_B Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLQ_C Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_A Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_B Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_C Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLS_A Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron] |
| 2NSP_A | 1.03e-12 | 997 | 1359 | 14 | 339 | ChainA, Pectinesterase A [Dickeya dadantii 3937],2NSP_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NST_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NST_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT6_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT6_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT9_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT9_B Chain B, Pectinesterase A [Dickeya dadantii 3937] |
| 4PMH_A | 3.90e-10 | 1151 | 1277 | 168 | 295 | Thestructure of rice weevil pectin methyl esterase [Sitophilus oryzae] |
| 5C1C_A | 1.28e-09 | 1149 | 1323 | 105 | 270 | CrystalStructure of the Pectin Methylesterase from Aspergillus niger in Deglycosylated Form [Aspergillus niger ATCC 1015] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P22751 | 8.36e-59 | 1849 | 2491 | 81 | 646 | Pectate disaccharide-lyase OS=Dickeya chrysanthemi OX=556 GN=pelX PE=1 SV=1 |
| P0C1A6 | 6.67e-40 | 2202 | 2494 | 40 | 326 | Pectate lyase L OS=Dickeya chrysanthemi OX=556 GN=pelL PE=3 SV=1 |
| P0C1A7 | 1.36e-38 | 2202 | 2494 | 40 | 326 | Pectate lyase L OS=Dickeya dadantii (strain 3937) OX=198628 GN=pelL PE=1 SV=1 |
| Q9ZQA3 | 1.67e-11 | 1086 | 1284 | 117 | 318 | Probable pectinesterase 15 OS=Arabidopsis thaliana OX=3702 GN=PME15 PE=2 SV=1 |
| Q8VYZ3 | 1.39e-10 | 1000 | 1339 | 95 | 372 | Probable pectinesterase 53 OS=Arabidopsis thaliana OX=3702 GN=PME53 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
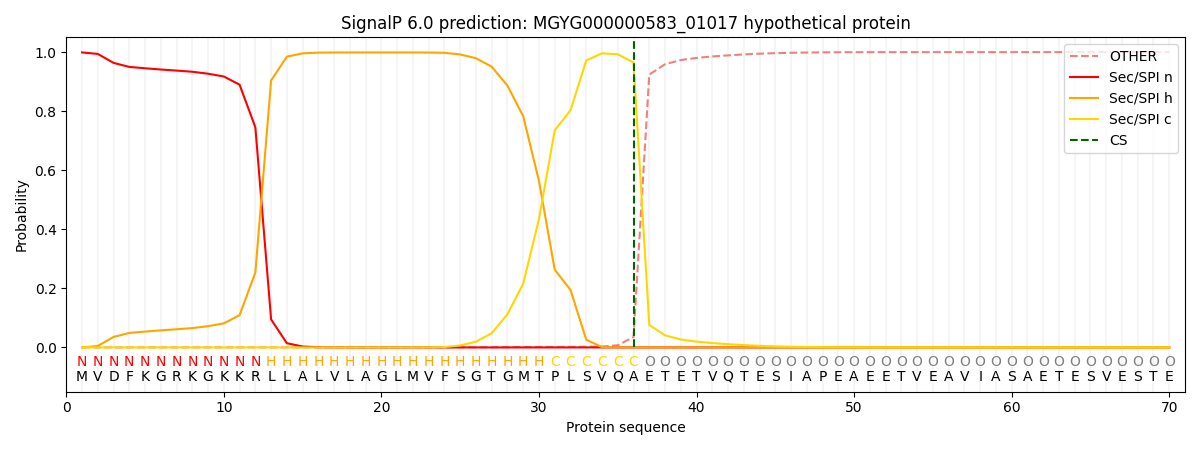
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000644 | 0.997099 | 0.001624 | 0.000230 | 0.000192 | 0.000166 |