You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000584_00836
You are here: Home > Sequence: MGYG000000584_00836
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | RC9 sp900544195 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; UBA932; RC9; RC9 sp900544195 | |||||||||||
| CAZyme ID | MGYG000000584_00836 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 30771; End: 32999 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH28 | 435 | 722 | 1.6e-33 | 0.7630769230769231 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5434 | Pgu1 | 1.30e-11 | 444 | 607 | 199 | 366 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| PLN03010 | PLN03010 | 6.69e-09 | 450 | 609 | 139 | 280 | polygalacturonase |
| pfam13229 | Beta_helix | 3.76e-07 | 480 | 614 | 3 | 141 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| PLN02793 | PLN02793 | 1.28e-05 | 452 | 614 | 145 | 304 | Probable polygalacturonase |
| PLN03003 | PLN03003 | 1.71e-05 | 451 | 607 | 114 | 259 | Probable polygalacturonase At3g15720 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBL02017.1 | 1.03e-131 | 40 | 731 | 39 | 734 |
| QRP58902.1 | 9.38e-129 | 4 | 742 | 16 | 775 |
| QQA09745.1 | 9.38e-129 | 4 | 742 | 16 | 775 |
| QQT79214.1 | 9.38e-129 | 4 | 742 | 16 | 775 |
| ASW16403.1 | 9.38e-129 | 4 | 742 | 16 | 775 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A7PZL3 | 3.28e-08 | 444 | 627 | 168 | 345 | Probable polygalacturonase OS=Vitis vinifera OX=29760 GN=GSVIVT00026920001 PE=1 SV=1 |
| Q9LW07 | 1.50e-06 | 451 | 607 | 114 | 259 | Probable polygalacturonase At3g15720 OS=Arabidopsis thaliana OX=3702 GN=At3g15720 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
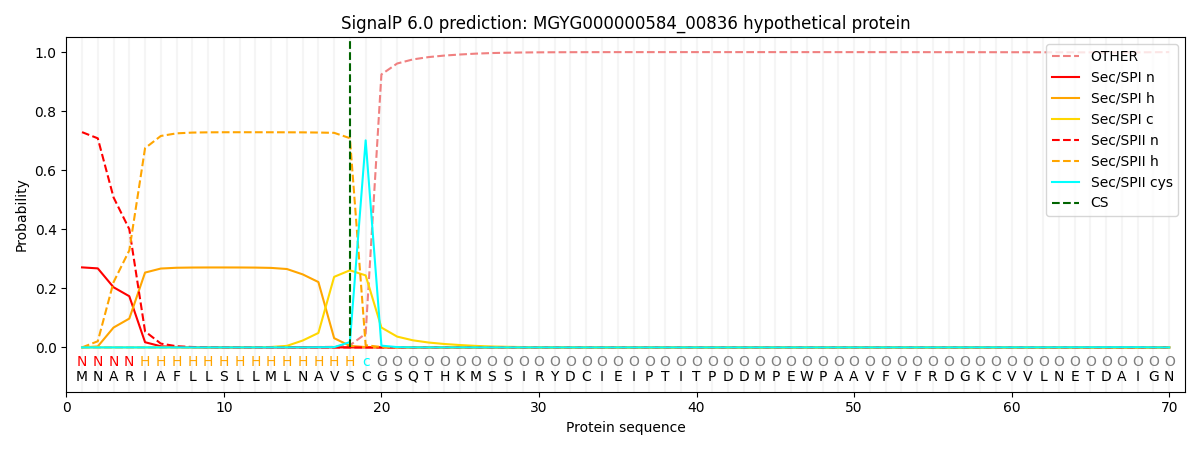
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000285 | 0.266297 | 0.733098 | 0.000110 | 0.000102 | 0.000092 |
