You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000584_00845
You are here: Home > Sequence: MGYG000000584_00845
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | RC9 sp900544195 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; UBA932; RC9; RC9 sp900544195 | |||||||||||
| CAZyme ID | MGYG000000584_00845 | |||||||||||
| CAZy Family | GH123 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 51041; End: 54985 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 731 | 997 | 4.7e-49 | 0.9768518518518519 |
| GH123 | 56 | 546 | 6.2e-42 | 0.8884758364312267 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00933 | Glyco_hydro_3 | 5.14e-25 | 683 | 996 | 9 | 280 | Glycosyl hydrolase family 3 N terminal domain. |
| pfam13320 | DUF4091 | 3.72e-20 | 465 | 530 | 3 | 66 | Domain of unknown function (DUF4091). This presumed domain is functionally uncharacterized. This domain family is found in bacteria, archaea and eukaryotes, and is approximately 70 amino acids in length. There is a single completely conserved residue G that may be functionally important. |
| PRK15098 | PRK15098 | 9.75e-19 | 762 | 1264 | 115 | 600 | beta-glucosidase BglX. |
| PLN03080 | PLN03080 | 1.03e-09 | 756 | 1301 | 103 | 634 | Probable beta-xylosidase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AFL77426.1 | 0.0 | 579 | 1312 | 9 | 743 |
| CBK63828.1 | 0.0 | 579 | 1312 | 9 | 743 |
| QIA06568.1 | 2.60e-314 | 579 | 1312 | 25 | 778 |
| QIX66430.1 | 2.32e-303 | 592 | 1312 | 40 | 775 |
| BBE20351.1 | 1.79e-302 | 586 | 1314 | 30 | 775 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5Z87_A | 1.18e-30 | 625 | 1301 | 30 | 669 | ChainA, EmGH1 [Aurantiacibacter marinus],5Z87_B Chain B, EmGH1 [Aurantiacibacter marinus] |
| 5M6G_A | 3.97e-21 | 716 | 1300 | 100 | 614 | Crystalstructure Glucan 1,4-beta-glucosidase from Saccharopolyspora erythraea [Saccharopolyspora erythraea D] |
| 5JP0_A | 5.91e-21 | 722 | 1095 | 84 | 417 | Bacteroidesovatus Xyloglucan PUL GH3B with bound glucose [Bacteroides ovatus],5JP0_B Bacteroides ovatus Xyloglucan PUL GH3B with bound glucose [Bacteroides ovatus] |
| 5Z9S_A | 9.53e-20 | 732 | 1301 | 83 | 664 | Functionaland Structural Characterization of a beta-Glucosidase Involved in Saponin Metabolism from Intestinal Bacteria [Bifidobacterium longum],5Z9S_B Functional and Structural Characterization of a beta-Glucosidase Involved in Saponin Metabolism from Intestinal Bacteria [Bifidobacterium longum] |
| 1LQ2_A | 3.36e-16 | 718 | 1080 | 65 | 394 | Crystalstructure of barley beta-D-glucan glucohydrolase isoenzyme Exo1 in complex with gluco-phenylimidazole [Hordeum vulgare],1X38_A crystal structure of barley beta-D-glucan glucohydrolase isoenzyme exo1 in complex with gluco-phenylimidazole [Hordeum vulgare],1X39_A Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme exo1 in complex with gluco-phenylimidazole [Hordeum vulgare] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q46684 | 2.29e-89 | 606 | 1298 | 33 | 652 | Periplasmic beta-glucosidase/beta-xylosidase OS=Dickeya chrysanthemi OX=556 GN=bgxA PE=3 SV=1 |
| B8NGU6 | 4.49e-88 | 631 | 1303 | 42 | 625 | Probable beta-glucosidase C OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=bglC PE=3 SV=1 |
| Q2UFP8 | 9.32e-88 | 616 | 1303 | 33 | 629 | Probable beta-glucosidase C OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=bglC PE=3 SV=2 |
| Q5BCC6 | 2.24e-85 | 627 | 1300 | 34 | 615 | Beta-glucosidase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=bglC PE=1 SV=1 |
| A7LXU3 | 3.31e-20 | 722 | 1095 | 106 | 439 | Beta-glucosidase BoGH3B OS=Bacteroides ovatus (strain ATCC 8483 / DSM 1896 / JCM 5824 / BCRC 10623 / CCUG 4943 / NCTC 11153) OX=411476 GN=BACOVA_02659 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
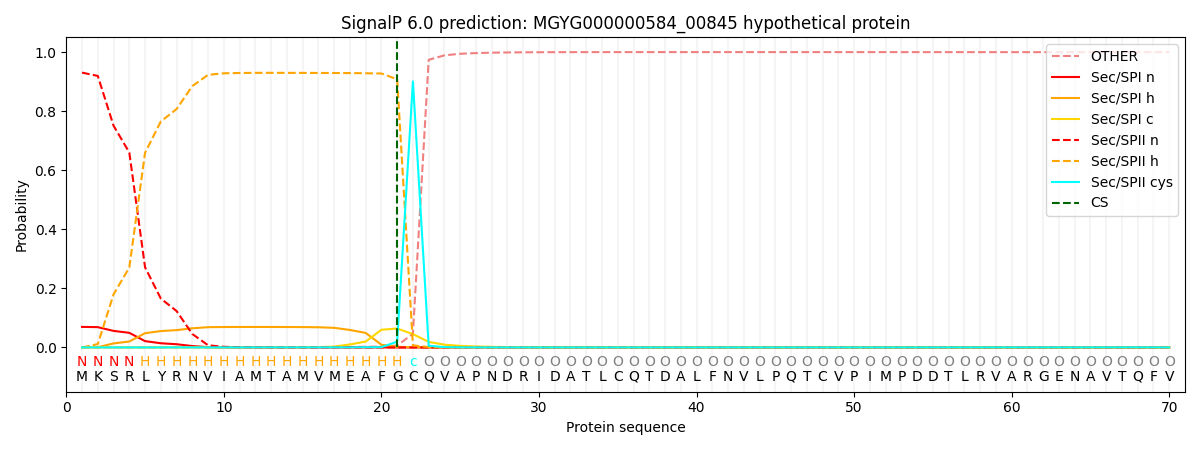
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000226 | 0.067164 | 0.932500 | 0.000043 | 0.000059 | 0.000037 |
