You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000589_01009
You are here: Home > Sequence: MGYG000000589_01009
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Faecalibacterium; | |||||||||||
| CAZyme ID | MGYG000000589_01009 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 891; End: 3593 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH28 | 90 | 412 | 4.8e-32 | 0.7569230769230769 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5434 | Pgu1 | 1.49e-32 | 61 | 422 | 79 | 416 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| pfam13229 | Beta_helix | 2.50e-06 | 231 | 343 | 48 | 154 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam12708 | Pectate_lyase_3 | 1.30e-05 | 67 | 324 | 4 | 210 | Pectate lyase superfamily protein. This family of proteins possesses a beta helical structure like Pectate lyase. This family is most closely related to glycosyl hydrolase family 28. |
| pfam10518 | TAT_signal | 2.57e-05 | 5 | 27 | 1 | 23 | TAT (twin-arginine translocation) pathway signal sequence. |
| pfam13229 | Beta_helix | 0.001 | 231 | 394 | 2 | 154 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AXB28928.1 | 0.0 | 1 | 900 | 19 | 918 |
| CBL02329.1 | 0.0 | 1 | 900 | 1 | 900 |
| BCJ93914.1 | 2.60e-295 | 45 | 899 | 4 | 882 |
| QOY86475.1 | 3.00e-93 | 58 | 712 | 17 | 492 |
| AHG93184.1 | 6.37e-92 | 62 | 712 | 32 | 507 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5OLP_A | 3.98e-13 | 57 | 335 | 37 | 315 | Galacturonidase[Bacteroides thetaiotaomicron VPI-5482],5OLP_B Galacturonidase [Bacteroides thetaiotaomicron VPI-5482] |
| 4MXN_A | 6.52e-09 | 65 | 293 | 22 | 215 | Crystalstructure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184],4MXN_B Crystal structure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184],4MXN_C Crystal structure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184],4MXN_D Crystal structure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as TAT
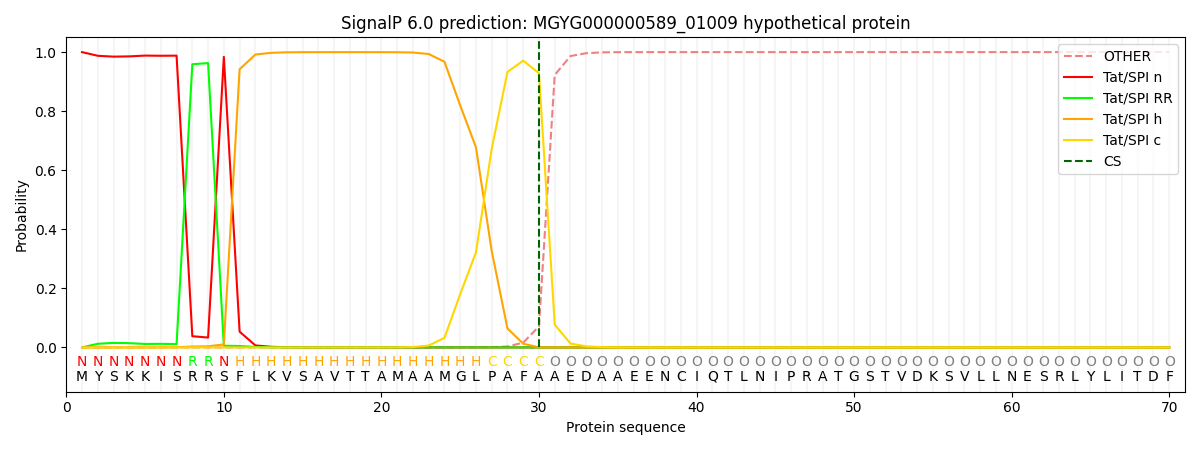
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 0.000000 | 0.999964 | 0.000000 | 0.000000 |
