You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000592_01248
You are here: Home > Sequence: MGYG000000592_01248
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; UBA932; RC9; | |||||||||||
| CAZyme ID | MGYG000000592_01248 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 883; End: 4179 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 350 | 1038 | 3.6e-79 | 0.6808510638297872 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 2.70e-24 | 377 | 817 | 30 | 423 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| pfam00754 | F5_F8_type_C | 3.29e-09 | 244 | 335 | 22 | 125 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| PRK10150 | PRK10150 | 5.93e-06 | 415 | 753 | 66 | 352 | beta-D-glucuronidase; Provisional |
| PRK10340 | ebgA | 6.39e-06 | 436 | 799 | 124 | 449 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| pfam02837 | Glyco_hydro_2_N | 8.06e-05 | 417 | 491 | 68 | 135 | Glycosyl hydrolases family 2, sugar binding domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities and has a jelly-roll fold. The domain binds the sugar moiety during the sugar-hydrolysis reaction. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRO23425.1 | 0.0 | 24 | 1084 | 60 | 1102 |
| ALO48125.1 | 0.0 | 24 | 1084 | 48 | 1082 |
| QUT49978.1 | 0.0 | 29 | 1084 | 71 | 1112 |
| BAR52607.1 | 0.0 | 29 | 1084 | 73 | 1113 |
| AEW22509.1 | 0.0 | 29 | 1084 | 73 | 1113 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5N6U_A | 3.21e-39 | 373 | 996 | 40 | 669 | Crystalstructure of Beta-D-Mannosidase from Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12],5N6U_B Crystal structure of Beta-D-Mannosidase from Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12],5N6U_C Crystal structure of Beta-D-Mannosidase from Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12],5N6U_D Crystal structure of Beta-D-Mannosidase from Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12] |
| 6BYE_A | 2.55e-38 | 374 | 968 | 30 | 658 | Crystalstructure of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri in complex with mannose [Xanthomonas citri pv. citri str. 306],6BYE_B Crystal structure of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri in complex with mannose [Xanthomonas citri pv. citri str. 306] |
| 6BYC_A | 2.56e-38 | 374 | 968 | 30 | 658 | Crystalstructure of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306] |
| 6BYI_A | 5.86e-38 | 374 | 968 | 28 | 656 | Crystalstructure of the acid-base mutant (E477A) of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306],6BYI_B Crystal structure of the acid-base mutant (E477A) of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306] |
| 6BYG_A | 1.36e-37 | 374 | 968 | 30 | 658 | Crystalstructure of the nucleophile mutant (E575A) of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306],6BYG_B Crystal structure of the nucleophile mutant (E575A) of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q75W54 | 4.79e-75 | 351 | 1084 | 13 | 791 | Mannosylglycoprotein endo-beta-mannosidase OS=Arabidopsis thaliana OX=3702 GN=EBM PE=1 SV=3 |
| Q5H7P5 | 8.19e-72 | 351 | 1022 | 10 | 731 | Mannosylglycoprotein endo-beta-mannosidase OS=Lilium longiflorum OX=4690 GN=EBM PE=1 SV=4 |
| Q4R1C4 | 2.92e-38 | 349 | 1069 | 36 | 763 | Exo-beta-D-glucosaminidase OS=Hypocrea jecorina OX=51453 GN=gls93 PE=1 SV=1 |
| C0LRA7 | 6.72e-38 | 349 | 1069 | 34 | 761 | Exo-beta-D-glucosaminidase OS=Hypocrea virens OX=29875 GN=gls1 PE=2 SV=1 |
| D4AUH1 | 7.83e-33 | 349 | 1069 | 39 | 761 | Exo-beta-D-glucosaminidase ARB_07888 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_07888 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
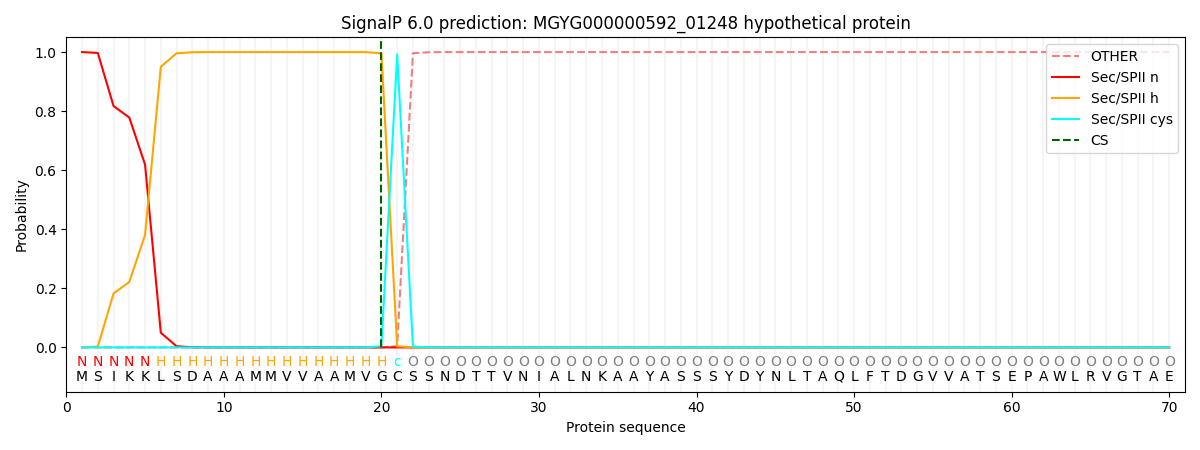
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000001 | 1.000050 | 0.000000 | 0.000000 | 0.000000 |
