You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000601_01055
You are here: Home > Sequence: MGYG000000601_01055
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
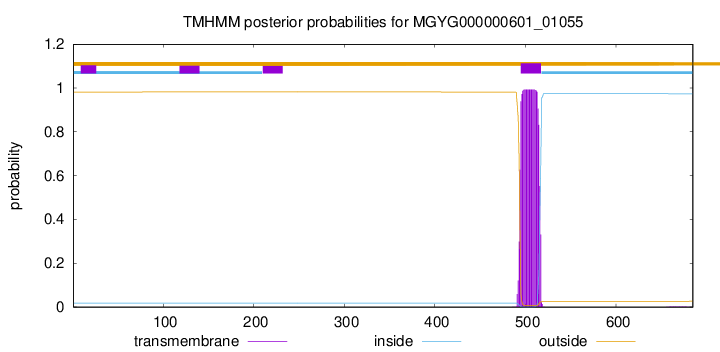
TMHMM annotations
Basic Information help
| Species | Mobilibacterium timonense | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Peptostreptococcales; Anaerovoracaceae; Mobilibacterium; Mobilibacterium timonense | |||||||||||
| CAZyme ID | MGYG000000601_01055 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 205; End: 2262 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT2 | 244 | 371 | 1.9e-20 | 0.7705882352941177 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| TIGR04283 | glyco_like_mftF | 8.03e-63 | 243 | 435 | 1 | 189 | transferase 2, rSAM/selenodomain-associated. This enzyme may transfer a nucleotide, or it sugar moiety, as part of a biosynthetic pathway. Other proposed members of the pathway include another transferase (TIGR04282), a phosphoesterase, and a radical SAM enzyme (TIGR04167) whose C-terminal domain (pfam12345) frequently contains a selenocysteine. [Unknown function, Enzymes of unknown specificity] |
| pfam02397 | Bac_transf | 5.24e-62 | 489 | 681 | 1 | 179 | Bacterial sugar transferase. This Pfam family represents a conserved region from a number of different bacterial sugar transferases, involved in diverse biosynthesis pathways. |
| TIGR03025 | EPS_sugtrans | 7.48e-60 | 484 | 681 | 253 | 438 | exopolysaccharide biosynthesis polyprenyl glycosylphosphotransferase. Members of this family are generally found near other genes involved in the biosynthesis of a variety of exopolysaccharides. These proteins consist of two fused domains, an N-terminal hydrophobic domain of generally low conservation and a highly conserved C-terminal sugar transferase domain (pfam02397). Characterized and partially characterized members of this subfamily include Salmonella WbaP (originally RfbP), E. coli WcaJ, Methylobacillus EpsB, Xanthomonas GumD, Vibrio CpsA, Erwinia AmsG, Group B Streptococcus CpsE (originally CpsD), and Streptococcus suis Cps2E. Each of these is believed to act in transferring the sugar from, for instance, UDP-glucose or UDP-galactose, to a lipid carrier such as undecaprenyl phosphate as the first (priming) step in the synthesis of an oligosaccharide "block". This function is encoded in the C-terminal domain. The liposaccharide is believed to be subsequently transferred through a "flippase" function from the cytoplasmic to the periplasmic face of the inner membrane by the N-terminal domain. Certain closely related transferase enzymes, such as Sinorhizobium ExoY and Lactococcus EpsD, lack the N-terminal domain and are not found by this model. |
| TIGR03022 | WbaP_sugtrans | 4.64e-57 | 486 | 681 | 256 | 449 | Undecaprenyl-phosphate galactose phosphotransferase, WbaP. The WbaP (formerly RfbP) protein has been characterized as the first enzyme in O-antigen biosynthesis in Salmonella typhimurium. The enzyme transfers galactose from UDP-galactose to a polyprenyl carrier (utilizing the highly conserved C-terminal sugar transferase domain, pfam02397) a reaction which takes place at the cytoplasmic face of the inner membrane. The N-terminal hydrophobic domain is then believed to facilitate the "flippase" function of transferring the liposaccharide unit from the cytoplasmic face to the periplasmic face of the inner membrane. This model includes the enterobacterial enzymes, where the function is presumed to be identical to the S. typhimurium enzyme as well as a somewhat broader group which are likely to catalyze the same or highly similar reactions based on a phylogenetic tree-building analysis of the broader sugar transferase family. Most of these genes are found within large operons dedicated to the production of complex exopolysaccharides such as the enterobacterial O-antigen. The most likely heterogeneity would be in the precise nature of the sugar molecule transferred. |
| TIGR04282 | glyco_like_cofC | 9.98e-55 | 20 | 212 | 2 | 189 | transferase 1, rSAM/selenodomain-associated. Members of this protein family show strongly correlated phylogenetic distribution, and in most cases co-clustering, with an unusual radical SAM enzyme (TIGR04167) whose C-terminal pfam12345 domain often contains a selenocysteine residue. Other members of the conserved gene neighborhood include another putative glycosyltransferase, an alkylhydroperoxidase family protein (TIGR04169), and a phosphoesterase family protein (TIGR04168). The cassette is likely to be biosynthetic but its exact function is unknown. [Unknown function, Enzymes of unknown specificity] |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBK78105.1 | 3.53e-103 | 17 | 455 | 3 | 427 |
| AVM48486.1 | 2.64e-101 | 18 | 469 | 13 | 448 |
| SET56509.1 | 2.62e-99 | 18 | 463 | 4 | 436 |
| QRP38681.1 | 3.46e-99 | 18 | 455 | 4 | 426 |
| ASN96524.1 | 3.46e-99 | 18 | 455 | 4 | 426 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5W7L_A | 5.84e-21 | 487 | 683 | 9 | 186 | Structureof Campylobacter concisus PglC I57M/Q175M variant [Campylobacter concisus 13826],5W7L_B Structure of Campylobacter concisus PglC I57M/Q175M variant [Campylobacter concisus 13826] |
| 3CGX_A | 8.26e-15 | 20 | 222 | 8 | 208 | ChainA, Putative nucleotide-diphospho-sugar transferase [Oleidesulfovibrio alaskensis G20] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q57491 | 7.39e-33 | 489 | 674 | 275 | 459 | Uncharacterized sugar transferase HI_0872 OS=Haemophilus influenzae (strain ATCC 51907 / DSM 11121 / KW20 / Rd) OX=71421 GN=HI_0872 PE=3 SV=1 |
| P26406 | 1.95e-32 | 487 | 681 | 278 | 469 | Undecaprenyl-phosphate galactose phosphotransferase OS=Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) OX=99287 GN=rfbP PE=3 SV=2 |
| P14186 | 1.18e-31 | 488 | 678 | 28 | 218 | Exopolysaccharide production protein ExoY OS=Sinorhizobium fredii (strain NBRC 101917 / NGR234) OX=394 GN=exoY PE=3 SV=1 |
| Q02731 | 1.04e-30 | 488 | 678 | 28 | 218 | Exopolysaccharide production protein ExoY OS=Rhizobium meliloti (strain 1021) OX=266834 GN=exoY PE=3 SV=1 |
| Q46628 | 2.05e-28 | 487 | 674 | 279 | 465 | UDP-galactose-lipid carrier transferase OS=Erwinia amylovora OX=552 GN=amsG PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
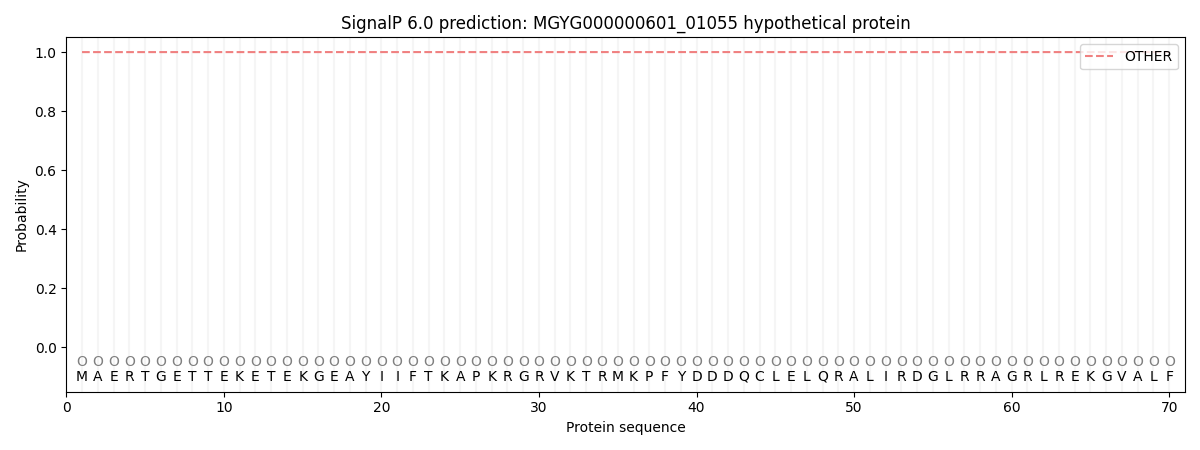
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000044 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |