You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000604_00197
You are here: Home > Sequence: MGYG000000604_00197
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UBA1829 sp900549415 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; UBA1829; UBA1829; UBA1829 sp900549415 | |||||||||||
| CAZyme ID | MGYG000000604_00197 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 234736; End: 236736 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 118 | 352 | 3.4e-36 | 0.8278388278388278 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3934 | COG3934 | 1.97e-24 | 40 | 560 | 3 | 517 | Endo-1,4-beta-mannosidase [Carbohydrate transport and metabolism]. |
| pfam02836 | Glyco_hydro_2_C | 5.77e-08 | 136 | 384 | 62 | 291 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| PRK10150 | PRK10150 | 7.67e-04 | 136 | 276 | 339 | 475 | beta-D-glucuronidase; Provisional |
| COG3250 | LacZ | 0.001 | 62 | 274 | 320 | 473 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10340 | ebgA | 0.009 | 176 | 209 | 417 | 450 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AHF94684.1 | 4.74e-125 | 56 | 661 | 1 | 628 |
| AMJ67494.1 | 8.48e-124 | 13 | 635 | 5 | 636 |
| AVM43652.1 | 1.76e-121 | 43 | 666 | 19 | 645 |
| PVY35568.1 | 1.69e-119 | 39 | 630 | 6 | 605 |
| SDT99127.1 | 4.29e-115 | 39 | 632 | 9 | 605 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7LR1_A | 6.12e-09 | 24 | 406 | 3 | 363 | ChainA, Glycosyl hydrolase BlGH5_18 [Bifidobacterium longum subsp. longum ATCC 55813],7LR1_B Chain B, Glycosyl hydrolase BlGH5_18 [Bifidobacterium longum subsp. longum ATCC 55813],7LR1_C Chain C, Glycosyl hydrolase BlGH5_18 [Bifidobacterium longum subsp. longum ATCC 55813],7LR1_D Chain D, Glycosyl hydrolase BlGH5_18 [Bifidobacterium longum subsp. longum ATCC 55813],7LR2_A Chain A, Glycosyl hydrolase BlGH5_18 [Bifidobacterium longum subsp. longum ATCC 55813],7LR2_B Chain B, Glycosyl hydrolase BlGH5_18 [Bifidobacterium longum subsp. longum ATCC 55813],7LR2_C Chain C, Glycosyl hydrolase BlGH5_18 [Bifidobacterium longum subsp. longum ATCC 55813],7LR2_D Chain D, Glycosyl hydrolase BlGH5_18 [Bifidobacterium longum subsp. longum ATCC 55813] |
| 7LR6_A | 3.28e-08 | 24 | 406 | 3 | 363 | ChainA, Glycosyl hydrolase BlGH5_18 [Bifidobacterium longum subsp. longum ATCC 55813],7LR6_B Chain B, Glycosyl hydrolase BlGH5_18 [Bifidobacterium longum subsp. longum ATCC 55813],7LR6_C Chain C, Glycosyl hydrolase BlGH5_18 [Bifidobacterium longum subsp. longum ATCC 55813],7LR6_D Chain D, Glycosyl hydrolase BlGH5_18 [Bifidobacterium longum subsp. longum ATCC 55813] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A1DBV1 | 7.94e-08 | 72 | 315 | 155 | 393 | Probable mannan endo-1,4-beta-mannosidase F OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=manF PE=3 SV=1 |
| Q5AZ53 | 1.58e-07 | 132 | 326 | 115 | 326 | Mannan endo-1,4-beta-mannosidase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=manC PE=1 SV=1 |
| B0Y9E7 | 9.41e-07 | 19 | 315 | 81 | 376 | Probable mannan endo-1,4-beta-mannosidase F OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=manF PE=3 SV=2 |
| Q4WBS1 | 9.41e-07 | 19 | 315 | 81 | 376 | Mannan endo-1,4-beta-mannosidase F OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=manF PE=1 SV=2 |
| B8NIV9 | 1.31e-06 | 72 | 315 | 160 | 398 | Probable mannan endo-1,4-beta-mannosidase F OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=manF PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
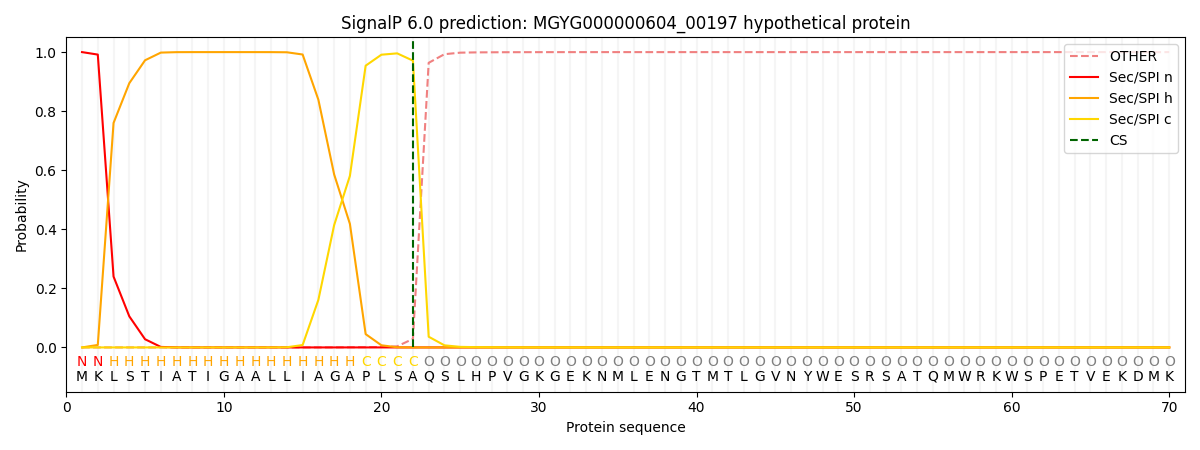
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000456 | 0.998679 | 0.000304 | 0.000188 | 0.000176 | 0.000166 |
